I’ve been excited about an idea for almost a year now, or more accurately maybe 6-7 months. I started thinking about it after developing the Flux Operator and realizing that we were not super great at running workflows in HPC-land. But then if we used Kubernetes (and yes, even on bare metal, which is an entire other project) we might be able to put together custom resource definitions (e.g., a Flux Framework MiniCluster) like LEGO to put together a workflow with many different kinds of steps. And the steps didn’t just have to be simulations or “HPC things” - they could be JobSet that combine services with applications in some interesting way, whether it needs MPI, GPUs, or what have you.
Background
My exploration started with workflow tools that are native to Kubernetes, so you can imagine I dipped into Argo and Kubeflow Pipelines and Flyte. To be frank, I didn’t have enough time to test these as much as I wanted, because I was just working on other things. I was frustrated with the refactor of Kubeflow Pipelines from version 1 to version 2 (where a lot of the functionality I needed was removed) but in retrospect they likely have (or had) plans to bring it back another way, and my timing was just off. But I think I was looking for something different. I didn’t want to have to tell scientists “Learn another, new workflow way” but rather “You can use the tools you’ve been using for a decade in this new environment.” I also wanted to provide seasoned workflow developers a strategy for writing executors (what they are typically called) to work not just with Kubernetes, but a set of converged environments (for HPC and cloud) in it. By the way, there are a ton of workflow tools!
The Queueing System: Kueue
I discovered Kueue a little later in the year, and it also took me a while to try out. It is (in simple terms) a job queueing system for Kubernetes. It doesn’t just hold the queue, however, it also manages resource groups and decides when a job should be admitted (the pods allowed to be created so a job can run) and when they should be deleted. If you have used high performance computing workload managers, this would correpond to the queue of jobs. In my mind, this approach mapped very nicely between the two spaces. We have a queue of things to run! And then I made the connection - if we can submit workflow tool steps to our traditional HPC queues, why not do the same but for Kubernetes?
The Workflow Tool: Snakemake
Thus my desire was to create some kind of simple prototype. I’ve developed for many workflow (or testing) tools, so it came down to a path of least resistance. I had been developing custom executors for Flux with Snakemake (1 and 2), but always was burdened by needing to add actual code to the Snakemake codebase. It was this final thing that encouraged me to push the lead developer of Snakemake, Johannes, to refactor the executor design to be modular, meaning that plugins would live in their own repositories and developers like myself would be empowered to develop and use them without an upstream contribution. This was quite a bit of work (and there is still more to be done and bugs to work out) but the early release is with Snakemake 8.0, and we now have the existing executors provided as modules thanks to the snakemake executor plugin interface! This gave me the setup that I needed for my prototype.
Converged Computing
While you may not traditionally think of Kubernetes as a place to run MPI, with the movement for converged computing, this is changing. Technologies like the Flux Operator and MPI Operator make it possible to run MPI workflows in Kubernetes. Since they are deployed as modular jobs (one or more pods working together) by an operator, this presents another opportunity for convergence - bringing together traditional workflow tools to submit not steps as jobs to an HPC system, but as operator custom resource definitions (CRD) to Kubernetes. This would allow simple steps to co-exist alongside steps that warrant more complex MPI. This is something I have been excited about for a while, and am (also) excited to share the first prototype here of that vision.
A Prototype
My goal was to design an executor plugin that would be able to generate custom resource definitions (ranging from a simple job to an MPIJob to a Flux MiniCluster job) submit with Kueue. The high level idea here is that Kueue is the cluster queue, again akin to an HPC queue. Yes, when we make things in Kubernetes we tend to change the first letter to a “K” that is just a thing. 😆️ And actually, within the Flux Operator we technically have another nested level of scheduler and queue (it’s hierarchical) for your workflow step, but that’s another point of discussion.
For the operating user, they would need to have Snakemake installed locally along with the plugin, and then access to a Kubernetes cluster with Kueue installed and queues configured. That’s the starting point. From there, we can take an already existing Snakefile, and (perhaps with some tweaks to tell the executor which step should use which operator) we can run our workflow. Actually, I’d go as far as to say with an approach like this, we can likely design new kinds of workflows (with single steps that use both services and applications) that were not possible or easily possible before.
Snakemake would thus be our workflow management system. And note this idea can extend to others, as long as they are able to have custom executors (many do, and most that I’ve developed for). It’s important to distinguish “workload” from “workflow” here -
- A workflow is generally referring to the steps in a DAG, each a unit of work that we might call a job.
- A workload is that job or single step that is handed to the workload manager
The workload manager doesn’t know anything about logic of steps - it receives modular pieces of work or tasks, and ensures that resources are assigned and the work gets done! On the other hand, Snakemake (as the workflow tool) is not concerned with a queue of work, but rather preparing steps from a directed acyclic graph (DAG) and then submitting the steps as jobs to a workload manager. The two do work together because each workflow step knows the resources it needs, and tells the workload manager to request them properly. Traditionally, many successful workflow tools have been developed for the biosciences, meaning that individual steps come down to running tools like bwa or samtools, and with little integration of high performance computing technologies like MPI. Let’s talk about what this might look like with a simple example, below.
The Workflow Tool
In the above, we start with a workflow tool. In this case we are using Snakemake. The workflow tool is able to take a specification file, which in this case is the Snakefile, a human understandable definition of a workflow, and convert it into a directed acyclic graph, or DAG, which is essentially a directed graph. In this graph, each step can be thought of as a single job in the workflow that will receive it’s own inputs, environment, and even container (especially in the case of Kubernetes) and then is expected to produce some output files. This is shown in the diagram as the green box on the left feeding into the blue DAG on the right.
The Workflow Steps (Jobs)
The modularity of a DAG also makes it amenable to operators. For example, if we have a step that runs LAMMPS simulations and needs MPI, we might submit a step to the Flux Operator to run a Flux Framework cluster in Kubernetes. If we just need to run a bash script for some analysis and don’t need that complexity, we might choose a job instead. To go back to our picture, we see that the DAG generated for this faux workflow has 5 steps, and each of them is given (at different times, represented by the length of the red lines that point to operator custom resource definitions generated) to Kubernetes.
Kubernetes
The core of Snakemake knows how to generate the DAG and time things, and the executor plugin is just given the specification for the job, and converts this into the CRDs. Each of these steps is going to be given (by Snakemake) to our queueing software, which in this case is Kueue. The snakemake kueue executor knows how to read the Snakefile and see what CRDs are desired for each step, and then prepare those custom resource definitions (yaml definitions). This is abstractly represented by the blue lines, which is where Kueue is taking a a step and providing a custom resource definition to a Kubernetes operator to complete it.
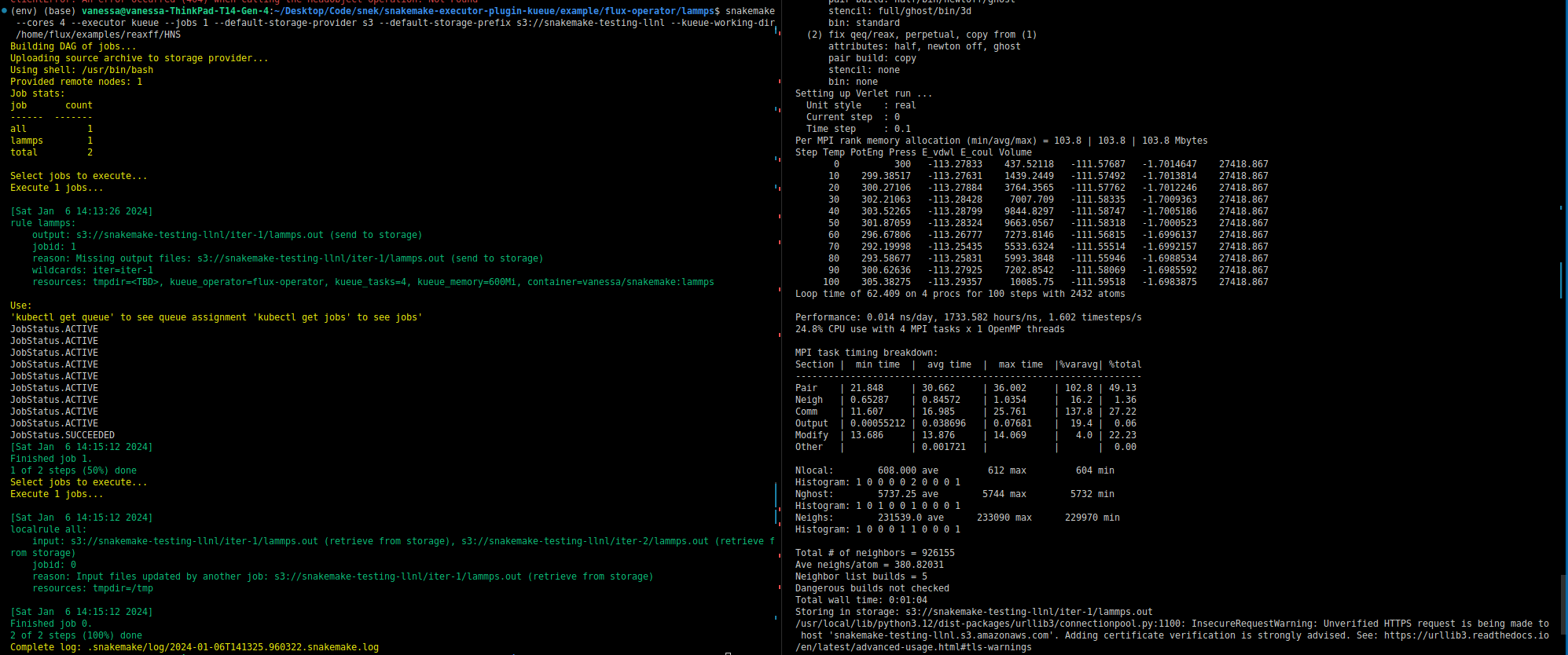
Importantly (to state this again) Kueue is not concerned with inputs and outputs. It is still the workflow software (Snakemake) that manages timing of things and inputs and outputs. If you were to look at the commands that the containers are running inside each operator, you’d see Snakemake logic to handle retrieval and saving of these artifacts. Snakemake will also know something is wrong if something is missing or not generated. For example, Snakemake will be looking for the input for step 2 from step 1, and will throw an error if it’s not there. Speaking of inputs and outputs, for this kind of setup where there isn’t a shared filesystem, the common strategy in bioinformatics is to use object or remote storage, and this is also built into Snakemake and represented in the picture as the gold object storage box that is accessed by all steps. When all is said and done, Snakemke is creating jobs to run in Kubernetes that know how to find their inputs and send back their outputs, and the Snakemake Kueue executor here orchestrates the entire thing! Just for funsies, here is the first successful run of LAMMPS (which uses MPI) in this environment.
Yay! That took me a hot minute to get working today. 😅️
Summary
The above is just in early development - I haven’t even finished the MPI Operator yet! The biggest challenge in my development has been working with the newly refactored Snakemake, which is asking for a Python version that is really new (much newer than many base images need) and then having different modules that are still a little buggy. I’ve provided custom Dockerfile with the examples where I get around these issues. But I got a basic “hello world” job working along with hello world and lammps running in the Flux Operator, and that alone has passed my threshold of “This idea has feet.” 🐾️
I like sharing things earlier than later, so I wanted to share it here. I hope to keep working on this in parallel to other things (it’s of interest to my group but not any kind of explicit focus, more of a fun project I’ve been working on in free time) so if it’s of interest to you, please reach out! I hope to make progress and maybe present it somewhere eventually so other workflow tool developers can try a similar approach. You can see the project here on GitHub if you are interested to try it out! MPI Operator support is prototyped and should be trivial to finish up if/when someone requests it.
Happy Saturday everyone!
Suggested Citation:
Sochat, Vanessa. "Snakemake Kueue Executor." @vsoch (blog), 06 Jan 2024, https://vsoch.github.io/2024/snakemake-kueue-executor/ (accessed 16 Apr 24).