wiki
Array Tomography Data Analysis Introduction
CAUTION: CURRENTLY BEING WRITTEN!
Setup
Here I am, I want to analyze AT data, and I haven’t a clue where to start. I will document my steps to provide future guidance to other weary graduate students.
Download data: in Gordon’s folder, the data is called “Test Dataset.” Normally the “images” come off of the microscope in special zipped files called zvis, from which we need to extract meta data and the image information. They then go through many stages of processing (artifact / blurry detection, alignment, stitching / registration, deconvolution) and ultimately wind up in the state that we see here… as a group of TIF files, one for each marker. Little did I know, TIF files can be 3D. When I dragged and dropped any “one” image into Fiji (ImageJ) I was surprised to see that it was in fact, hundreds.
Download scripts: in the lab Software Folder, the scripts are under the folder “Image Analysis Suite”
Add the path to MATLAB:
addpath(genpath('/home/vanessa/Documents/Code/MATLAB/AT/Image Analysis Suite'))
The only information that I had to start with was a diagram with the following workflow:
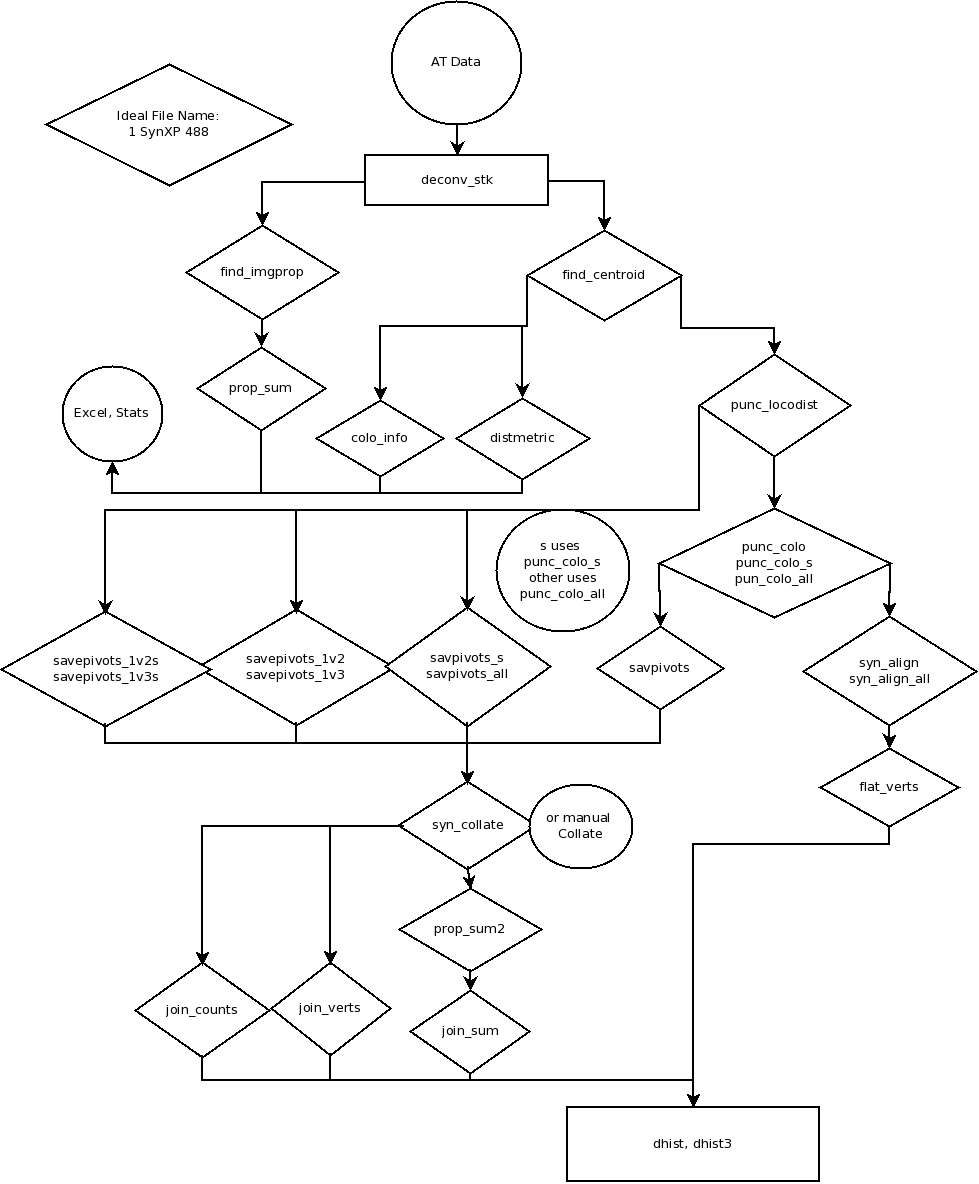
And the file “AT Functions List” provides a starting point for reviewing the scripts. I will include script descriptions from this file in my review.
Preparing Images
When I embarked on figuring this out, the scripts were beautifully documented, but it wasn’t clear exactly what constituted a “stack” of images to read into a horizontal cell array. A horizontal cell array of both image paths and full data have not yet worked. Update: The solution is to use a utility called “stack_gui” as follows: (Note that you can select multiple images)
[stk,range,filename,pathname,filterindex,info_out] = stack_gui(varargin)
It should be noted that the Test Dataset has already been deconvoluted (decon_stk) and so we can start with the “find_centroid.”
Label Centroids
1) Find centroid: Finds the centroids of punctas from an image stack as well as other pertinent data, such as brightness and size. Used as a starting point for punc_me_lite and punc_locodist.
USAGE:
[pivots_out,centroid_stk,lum_stk] = find_centroid(varargin)
[pivots,centroid_stk,lum_stk] = find_centroid('stks',array,'theshold',0.5,'type',2,'type2',[0 1],'norm',1)
‘stks’ is a horizontal cell array of the stacks. So we can start by going to the folder with our image TIFF files:
cd /home/vanessa/Documents/Work/AT/Test' Dataset'/
I had originally thought I’d need to read in the stacks on my own, and first I figured out that “stacks_gui” will do this for me (to create input for “find_centroid,” however I then learned that you can skip that all together and just call “find_centroid,” which will by default call “stacks_gui” if no input arguments are supplied.
[pivots_out,centroid_stk,lum_stk] = find_centroid
SCRIPT BUG LOG
- In find_centroid: If I specify a stk as an input argument, the channel_path variable is not set and it spits out an error on line 247 because it of course cannot make the directory.