If pulling a thread of meaning from woven text
is that which your heart does wish.
Not so absurd or seemingly complex,
if you befriend a tiny word fish.

I developed a simple tool for standard extraction of terminology and corpus, Wordfish, that is easily deployed to a cluster environment. I’m graduating (hopefully, tentatively, who knows) soon, and because publication is unlikely, I will write about the tool here, in the case it is useful to anyone. I did this project for fun, mostly because I found DeepDive to be overly complicated for my personal goal of extracting a simple set of terms from a corpus in the case that I couldn’t define relationships apriori (I wanted to learn them from the data). Thus I used neural networks (word2vec) to learn term relationships based on their context. I was able to predict reddit boards for different mental illness terms with high accuracy, and it sort of ended there because I couldn’t come up with a good application in Cognitive Neuroscience, and no “real” paper is going to write about predicting reddit boards. I was sad to not publish something, but then I realized I am empowered to write things on the internet. :) Not only that, I can make up my own rules. I don’t have to write robust methods with words, I will just show and link you to code. I might even just use bulletpoints instead of paragraphs. For results, I’ll link to ipython notebooks. I’m going to skip over the long prose and trust that if you want to look something up, you know how to use Google and Wikipedia. I will discuss the tool generally, and show an example of how it works. First, an aside about publication in general - feel free to skip this if you aren’t interested in discussing the squeaky academic machine.
Why sharing incomplete methods can be better than publication
It’s annoying that there is not a good avenue, or even more so, that it’s not desired or acceptable, to share a simple (maybe even incomplete) method or tool that could be useful to others in a different context. Publication requires the meaningful application. It’s annoying that, as researchers, we salivate for these “publication” things when the harsh reality is that this slow, inefficient process results in yet another PDF/printed thing with words on a page, offering some rosy description of an analysis and result (for which typically minimal code or data is shared) that makes claims that are over-extravagant in order to be sexy enough for publication in the first place (I’ve done quite a bit of this myself). A publication is a static thing that, at best, gets cited as evidence by another paper (and likely the person making the citation did not read the paper to full justice). Maybe it gets parsed from pubmed in someone’s meta analysis to try and “uncover” underlying signal across many publications that could have been transparently revealed in some data structure in the first place. Is this workflow really empowering others to collaboratively develop better methods and tools? I think not. Given the lack of transparency, I’m coming to realize that it’s much faster to just share things early. I don’t have a meaningful biological application. I don’t make claims that this is better than anything else. This is not peer reviewed by some three random people that gives it a blessing like from a rabbi. I understand the reasons for these things, but the process of conducting research, namely hiding code and results toward that golden nugget publication PDF, seems so against a vision of open science. Under this context, I present Wordfish.
Wordfish: tool for standard corpus and terminology extraction
Abstract
The extraction of entities and relationships between them from text is becoming common practice. The availability of numerous application program interfaces (API) to extract text from social networks, blogging platforms and feeds, standard sources of knowledge is continually expanding, offering an extensive and sometimes overwhelming source of data for the research scientist. While large corporations might have exclusive access to data and robust pipelines for easily obtaining the data, the individual researcher is typically granted limited access, and commonly must devote substantial amounts of time to writing extraction pipelines. Unfortunately, these pipelines are usually not extendable beyond the dissemination of any result, and the process is inefficiently repeated. Here I present Wordfish, a tiny but powerful tool for the extraction of corpus and terms from publicly available sources. Wordfish brings standardization to the definition and extraction of terminology sources, providing an open source repository for developers to write plugins to extend their specific terminologies and corpus to the framework, and research scientists an easy way to select from these corpus and terminologies to perform extractions and drive custom analysis pipelines. To demonstrate the utility of this tool, I use Wordfish in a common research framework: classification. I first build deep learning models to predict Reddit boards from post content with high accuracy. I hope that a tool like Wordfish can be extended to include substantial plugins, and can allow easier access to ample sources of textual content for the researcher, and a standard workflow for developers to add a new terminology or corpus source.
Introduction
While there is much talk of “big data,” when you peek over your shoulder and look at your colleague’s dataset, there is a pretty good chance that it is small or medium sized. When I wanted to extract terms and relationships from text, I went to DeepDive, the ultimate powerhouse to do this. However, I found that setting up a simple pipeline required database and programming expertise. I have this expertise, but it was tenuous. I thought that it should be easy to do some kind of NLP analysis, and combine across different corpus sources. When I started to think about it, we tend to reuse the same terminologies (eg, an ontology) and corpus (pubmed, reddit, wikipedia, etc), so why not implement an extraction once, and then provide that code for others? This general idea would make a strong distinction between a developer, meaning an individual best suited to write the extraction pipeline, and the researcher, an individual best suited to use it for analysis. This sets up the goals of Wordfish: to extract terms from a corpus, and then do some higher level analysis, and make it standardized and easy.
Wordfish includes data structures that can capture an input corpus or terminology, and provides methods for retrieval and extraction. Then, it allows researchers to create applications that interactively select from the available corpus and terminologies, deploy the applications in a cluster environment, and run an analysis. This basic workflow is possible and executable without needing to set up an entire infrastructure and re-writing the same extraction scripts that have been written a million times before.
Methods
The overall idea behind the infrastructure of wordfish is to provide terminologies, corpus, and an application for working with them in a modular fashion. This means that Wordfish includes two things, wordfish-plugins and wordfish-python. Wordfish plugins are modular folders, each of which provides a standard data structure to define extraction of a corpus, terminology or both. Wordfish python is a simple python module for generating an application, and then deploying the application on a server to run analyses.
Wordfish Plugins
A wordfish plugin is simply a folder with typically two things: a functions.py file to define functions for extraction, and a config.json that is read by wordfish-python to deploy the application. We can look at the structure of a typical plugin:
plugin functions.py init.py config.json
Specifically, the functions.py has the following functions:
1) extract_terms: function to call to return a data structure of terms
2) extract_text: function to call to return a data structure of text (corpus)
3) extract_relations: function to call to return a data structure of relations
4) functions.py: is the file in the plugin folder to store these functions
The requirement of every functions.py is an import of general functions from wordfish-python that will save a data structure for a corpus, terminology, or relationships:
# IMPORTS FOR ALL PLUGINS
from wordfish.corpus import save_sentences
from wordfish.terms import save_terms
from wordfish.terms import save_relations
from wordfish.plugin import generate_job
The second requirement is a function, go_fish, which is the main function to be called by wordfish-python under the hood. In this function, the user writing the plugin can make as many calls to generate_job as necessary. A call to generate job means that a slurm job file will be written to run a particular function (func) with a specified category or extraction type (e.g., terms, corpus, or relations). This second argument helps the application determine how to save the data. A go_fish function might look like this:
# REQUIRED WORDFISH FUNCTION
def go_fish():
generate_job(func="extract_terms",category="terms")
generate_job(func="extract_relations",category="relations")
The above will generate slurm job files to be run to extract terms and relations. Given input arguments are required for the function, the specification can look as follows:
generate_job(func="extract_relations",inputs={"terms":terms,"maps_dir":maps_dir},category="relations",batch_num=100)
where inputs is a dictionary of keys being variable names, values being the variable value. The addition of the batch_num variable also tells the application to split the extraction into a certain number of batches, corresponding to SLURM jobs. This is needed in the case that running a node on a cluster is limited to some amount of time, and the user wants to further parallelize the extraction.
Extract terms
Now we can look at more detail at the extract_terms function. For example, here is this function for the cognitive atlas. The extract_terms will return a json structure of terms
def extract_terms(output_dir):
terms = get_terms()
save_terms(terms,output_dir=output_dir)
You will notice that the extract_terms function uses another function that is defined in functions.py, get_terms. The user is free to include in the wordfish-plugin folder any number of additional files or functions that assist toward the extraction. Here is what get_terms looks like:
def get_terms():
terms = dict()
concepts = get_concepts()
for c in range(len(concepts)):
concept_id = concepts[c]["id"]
meta = {"name":concepts[c]["name"],
"definition":concepts[c]["definition_text"]}
terms[concept_id] = meta
return terms
This example is again from the Cognitive Atlas, and we are parsing cognitive ceoncepts into a dictionary of terms. For each cognitive concept, we are preparing a dictionary (JSON data structure) with fields name, and definition. We then put that into another dictionary terms with the key as the unique id. This unique id is important in that it will be used to link between term and relations definitions. You can assume that the other functions (e.g., get_concepts are defined in the functions.py file.
Extract relations
For extract_relations we return a tuple of the format (term1_id,term2_id,relationship):
def extract_relations(output_dir):
links = []
terms = get_terms()
concepts = get_concepts()
for concept in concepts:
if "relationships" in concept:
for relation in concept["relationships"]:
relationship = "%s,%s" %(relation["direction"],relation["relationship"])
tup = (concept["id"],relation["id"],relationship)
links.append(tup)
save_relations(terms,output_dir=output_dir,relationships=links)
Extract text
Finally, extract_text returns a data structure with some unique id and a blob of text. Wordfish will parse and clean up the text. The data structure for a single article is again, just JSON:
corpus[unique_id] = {"text":text,"labels":labels}
Fields include the actual text, and any associated labels that are important for classification later. The corpus (a dictionary of these data structures) gets passed to save_sentences
save_sentences(corpus_input,output_dir=output_dir)
More detail is provided in the wordfish-plugin README
The plugin controller: config.json
The plugin is understood by the application by way of a folder’s config.json, which might look like the following:
[
{
"name": "NeuroSynth Database",
"tag": "neurosynth",
"corpus": "True",
"terms": "True",
"labels": "True",
"relationships":"True",
"dependencies": {
"python": [
"neurosynth",
"pandas"
],
"plugins": ["pubmed"]
},
"arguments": {
"corpus":"email"
},
"contributors": ["Vanessa Sochat"],
"doi": "10.1038/nmeth.1635",
}
]
1) name: a human readable description of the plugin
2) tag: a term (no spaces or special characters) that corresponds with the folder name in the plugins directory. This is a unique id for the plugin.
3) corpus/terms/relationships: boolean, each “True” or “False” should indicate if the plugin can return a corpus (text to be parsed by wordfish) or terms (a vocabulary to be used to find mentions of things), or relations (relationships between terms). This is used to parse current plugins available for each purpose, to show to the user.
4) dependencies: should include “python” and “plugins.” Python corresponds to python packages that are dependencies, and these plugins are installed by the overall application. Plugins refers to other plugins that are required, such as pubmed. This is an example of a plugin that does not offer to extract a specific corpus, terminology, or relations, but can be included in an application for other plugins to use. In the example above, the neurosynth plugin requires retrieving articles from pubmed, so the plugin develop specifies needing pubmed as a plugin dependency.
5) arguments: a dictionary with (optionally) corpus and/or terms. The user will be asked for these arguments to run the extract_text and extract_terms functions.
6) contributors: a name/orcid ID or email of researchers responsible for creation of the plugins. This is for future help and debugging.
7) doi: a reference or publication associated with the resource. Needed if it’s important for the creator of the plugin to ascribe credit to some published work.
Best practices for writing plugins
Given that a developer is writing a plugin, it is generally good practice to print to the screen what is going on, and how long it might take, as a courtesy to the user, if something needs review or debugging.
“Extracting relationships, will take approximately 3 minutes”
The developer should also use clear variable names, well documented and spaced functions (one liners are great in python, but it’s more understandable by the reader if to write out a loop sometimes), and attribute function to code that is not his. Generally, the developer should just follow good practice as a coder and human being.
Functions provided by Wordfish
While most users and clusters have internet connectivity, it cannot be assumed, and an error in attempting to access an online resource could trigger an error. If a plugin has functions that require connectivity, Wordfish provides a function to check:
from wordfish.utils import has_internet_connectivity
if has_internet_connectivity():
# Do analysis
If the developer needs a github repo, Wordfish has a function for that:
from wordfish.vm import download_repo
repo_directory = download_repo(repo_url="https://github.com/neurosynth/neurosynth-data")
If the developer needs a general temporary place to put things, tempfile is recommended:
import tempfile
tmpdir = tempfile.mkdtemp()
Wordfish has other useful functions for downloading data, or obtaining a url. For example:
from wordfish.utils import get_url, get_json
from wordfish.standards.xml.functions import get_xml_url
myjson = get_json(url)
webpage = get_url(url)
xml = get_xml_url(url)
Custom Applications with Wordfish Python
The controller, wordfish-python is a flask application that provides the user (who is just wanting to generate an application) with an interactive web interface for doing so. It is summarized nicely in the README:
Choose your input corpus, terminologies, and deployment environment, and an application will be generated to use deep learning to extract features for text, and then entities can be mapped onto those features to discover relationships and classify new texty things. Custom plugins will allow for dynamic generation of corpus and terminologies from data structures and standards of choice from wordfish-plugins You can have experience with coding (and use the functions in the module as you wish), or no experience at all, and let the interactive web interface walk you through generation of your application.
Installation can be done via github or pip:
pip install git+git://github.com/word-fish/wordfish-python.git
pip install wordfish
And then the tool is called to open up a web interface to generate the application:
wordfish
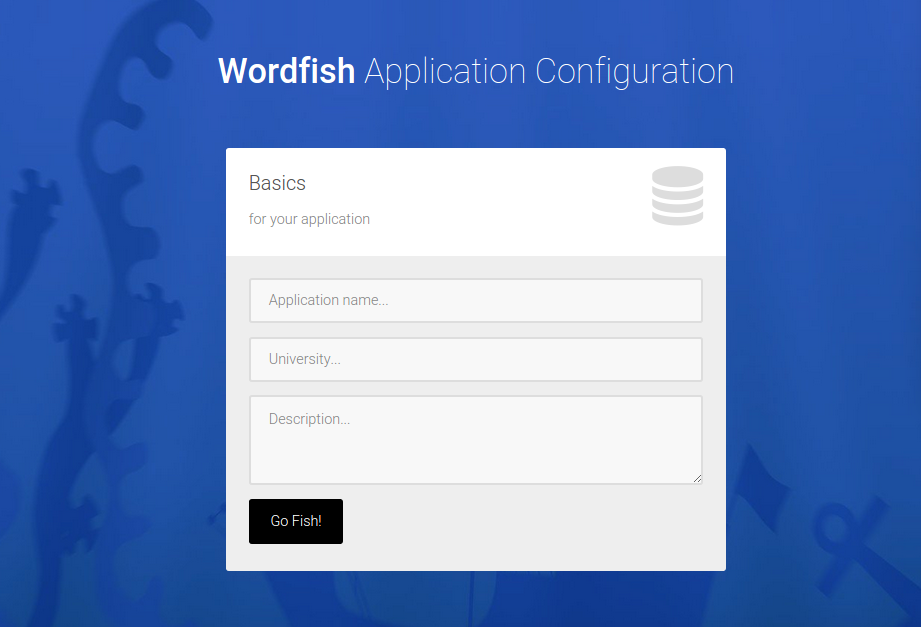
The user then selects terminologies and corpus.
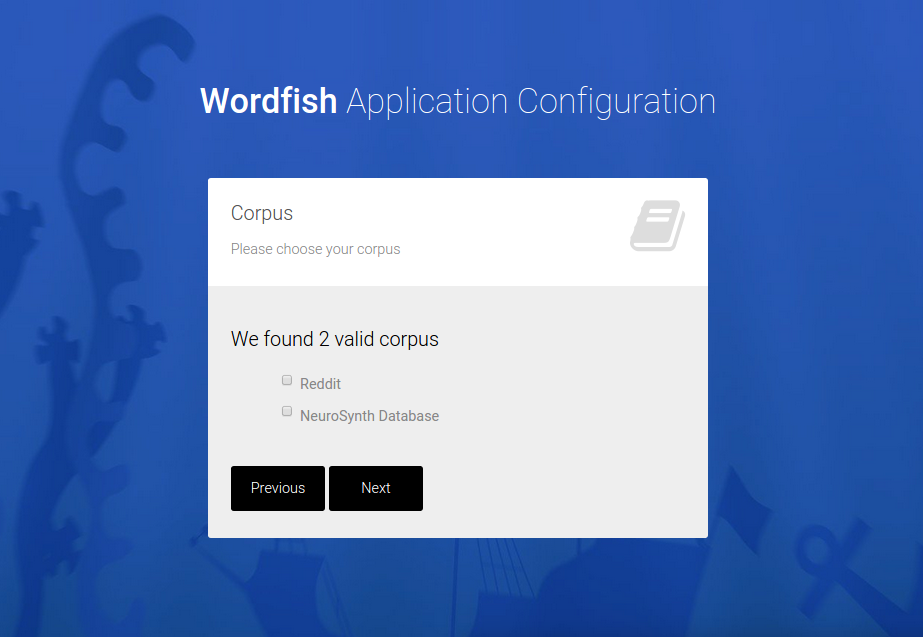
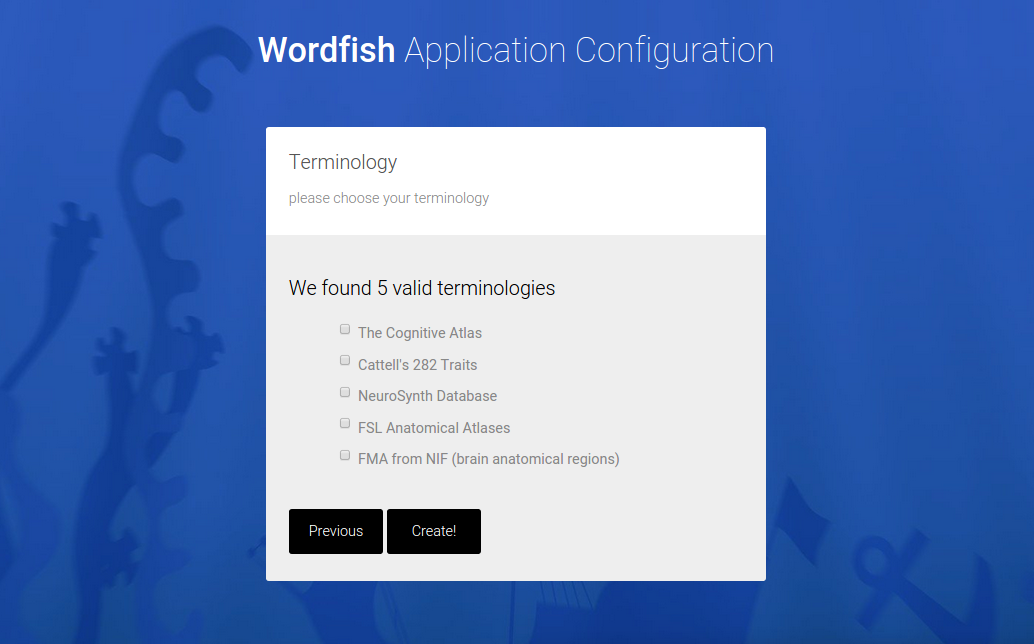
And a custom application is generated, downloaded as a zip file in the browser. A “custom application” means a folder that can be dropped into a cluster environment, and run to generate the analysis,
Installing in your cluster environment
The user can drop the folder into a home directory of the cluster environment, and run the install script to install the package itself, and generate the output folder structure. The only argument that is needed is to supply is the base of the output directory:
WORK=/scratch/users/vsochat/wordfish
bash install.sh $WORK
All scripts for the user to run are in the scripts folder here:
cd $WORK/scripts
Each of these files corresponds to a step in the pipeline, and is simply a list of commands to be run in parallel. The user can use launch, or submit each command to a SLURM cluster. A basic script is provided to help submit jobs to a SLURM cluster, and this could be tweaked to work with other clusters (e.g., SGE).
Running the Pipeline
After the installation of the custom application is complete, this install script simply runs run.py, which generates all output folders and running scripts. the user has a few options for running:
1) submit the commands in serial, locally. The user can run a job file with bash, bash run_extraction_relationships.job
2) submit the commands to a launch cluster, something like launch -s run_extraction_relationships.job
3) submit the commands individually to a slurm cluster. This will mean reading in the file, and submitting each script with a line like sbatch -p normal -j myjob.job [command line here]
Output structure
The jobs are going to generate output to fill in the following file structure in the project base folder, which again is defined as an environmental variable when the application is installed (files that will eventually be produced are shown):
WORK
corpus
corpus1
12345_sentences.txt
12346_sentences.txt
corpus2
12345_sentences.txt
12346_sentences.txt
terms
terms1_terms.txt
terms2_relationships.txt
scripts
run_extraction_corpus.job
run_extraction_relationships.job
run_extraction_terms.job
The folders are generated dynamically by the run.py script for each corpus and terms plugin based on the tag variable in the plugin’s config.json. Relationships, by way of being associated with terms, are stored in the equivalent folder, and the process is only separate because it is not the case that all plugins for terms can have relationships defined. The corpus are kept separate at this step as the output has not been parsed into any standard unique id space. Wordfish currently does not do this, but if more sophisticated applications are desired (for example with a relational database), this would be a good strategy to take.
Analysis
Once the user has files for corpus and terms, he could arguably do whatever he pleases with them. However, I found the word2vec neural network to be incredibly easy and cool, and have provided a simple analysis pipeline to use it. This example will merge all terms and corpus into a common framework, and then show examples of how to do basic comparisons, and vector extraction (custom analyses scripts can be based off of this). We will do the following:
1) Merge terms and relationships into a common corpus
2) For all text extract features with deep learning (word2vec)
3) Build classifiers to predict labels from vectors
Word2Vec Algorithm
First, what is a word2vec model? Generally, Word2Vec is a neural network implementation that will allow us to learn word embeddings from text, specifically a matrix of words by some N features that best predict the neighboring words for each term. This is an interesting way to model a text corpus because it’s not about occurrence, but rather context, of words, and we can do something like compare a term “anxiety” in different contexts. If you want equations, see this paper.
The problem Wordfish solves
Wordfish currently implements Word2Vec. Word2Vec is an unsupervised model. Applications like DeepDive take the approach that a researcher knows what he or she is looking for, requiring definition of entities as first step before their extraction from a corpus. This is not ideal given that a researcher has no idea about these relationships, or lacks either positive or negative training examples. In terms of computational requirements, Deepdive also has some that are unrealistic. For example, using the Stanford Parser is required to determine parts of speech and perform named entity recognition. While this approach is suitable for a large scale operation to mine very specific relationships between well-defined entities in a corpus, for the single researcher that wants to do simpler natural language processing, and perhaps doesn’t know what kind of relationships or entities to look for, it is too much. This researcher may want to search for some terms of interest across a few social media sources, and build models to predict one type of text content from another. The researcher may want to extract relationships between terms without having a good sense of what they are to begin with, and definition of entities, relationships, and then writing scripts to extract both should not be a requirement. While it is reasonable to ask modern day data scientists to partake in small amounts of programming, substantial setting up of databases and writing extraction pipelines should not be a requirement. A different approach that is taken by Wordfish is to provide plugins for the user to interactively select corpus and terminology, deploy their custom application in their computational environment of choice, and perform extraction using the tools that are part of their normal workflows, which might be a local command line or computing cluster.
When the DeepDive approach makes sense, the reality is that setting up the infrastructure to deploy DeepDive is really hard. When we think about it, the two applications are solving entirely different problems. All we really want to do is discover how terms are related in text. We can probably do ok to give DeepDive a list of terms, but then to have to “know” something about the features we want to extract, and have positive and negative cases for training is really annoying. If it’s annoying for a very simple toy analysis (finding relationships between cognitive concepts) I can’t even imagine how that annoyingness will scale when there are multiple terminologies to consider, different relationships between the terms, and a complete lack of positive and negative examples to validate. This is why I created Wordfish, because I wanted an unsupervised approach that required minimal set up to get to the result. Let’s talk a little more about the history of Word2Vec from this paper.
The N-Gram Model
The N-gram model (I think) is a form of hidden Markov Model where we model the P(word) given the words that came before it. The authors note that N-gram models work very well for large data, but in the case of smaller datasets, more complex methods can make up for it. However, it follows logically that a more complex model on a large dataset gives us the best of all possible worlds. Thus, people started using neural networks for these models instead.
simple models trained on huge amounts of data outperform complex systems trained on less data.
The high level idea is that we are going to use neural networks to represent words as vectors, word “embeddings.” Training is done with stochastic gradient descent and backpropagation.
How do we assess the quality of word vectors?
Similar words tend to be close together, and given a high dimensional vector space, multiple representations/relationships can be learned between words (see top of page 4). We can also perform algebraic computations on vectors and discover cool relationships, for example, the vector for V(King) - V(Man) + V(Woman) is close to V(Queen). The most common metric to compare vectors seems to be cosine distance. The interesting thing about this approach reported here is that by combining individual word vectors we can easily represent phrases, and learn new interesting relationships.
Two different algorithm options
You can implement a continuous bag of words (CBOW) or skip-gram model:
1) CBOW: predicts the word given the context (other words)
2) skip-gram: predicts other words (context) given a word (this seems more useful for what we want to do)
They are kind of like inverses of one another, and the best way to show this is with a picture:

Discarding Frequent Words
The paper notes that having frequent words in text is not useful, and that during training, frequent words are discarded with a particular probability based on the frequency. They use this probability in a sampling procedure when choosing words to train on so the more frequent words are less likely to be chosen. For more details, see here, and search Google.
Building Word2Vec Models
First, we will train a simple word2vec model with different corpus. And to do this we can import functions from Wordfish, which is installed by the application we generated above.
from wordfish.analysis import (build_models, save_models, export_models_tsv,
load_models, extract_similarity_matrix,
export_vectors, featurize_to_corpus)
from wordfish.models import build_svm
from wordfish.corpus import get_corpus, get_meta, subset_corpus
from wordfish.terms import get_terms
from wordfish.utils import mkdir
import os
Installation of the application also write the environmental variable WORDFISH_HOME to your bash profile, so we can reference it easily:
base_dir = os.environ["WORDFISH_HOME"]
It is generally good practice to keep all components of an analysis well organized in the output directory. It makes sense to store analyses, models, and vectors:
# Setup analysis output directories
analysis_dir = mkdir("%s/analysis" %(base_dir))
model_dir = mkdir("%s/models" %(analysis_dir))
vector_dir = mkdir("%s/vectors" %(analysis_dir))
Wordfish then has nice functions for generating a corpus, meaning removing stop words, excess punctuation, and the typical steps in NLP analyses. The function get_corpus returns a dictionary, with the key being the unique id of the corpus (the folder name, tag of the original plugin). We can then use the subset_corpus plugin if we want to split the corpus into the different groups (defined by the labels we specified in the initial data structure):
# Generate more specific corpus by way of file naming scheme
corpus = get_corpus(base_dir)
reddit = corpus["reddit"]
disorders = subset_corpus(reddit)
corpus.update(disorders)
We can then train corpus-specific models, meaning word2vec models.
# Train corpus specific models
models = build_models(corpus)
Finally, we can export models to tsv, export vectors, and save the model so we can easily load again.
# Export models to tsv, export vectors, and save
save_models(models,base_dir)
export_models_tsv(models,base_dir)
export_vectors(models,output_dir=vector_dir)
I want to note that I used gensim for learning and some methods. The work and examples from Dato are great!
Working with models
Wordfish provides functions for easily loading a model that is generated from a corpus:
model = load_models(base_dir)["neurosynth"]
You can then do simple things, like find the most similar words for a query word:
model.most_similar("anxiety")
# [('aggression', 0.77308839559555054),
# ('stress', 0.74644440412521362),
# ('personality', 0.73549789190292358),
# ('excessive', 0.73344630002975464),
# ('anhedonia', 0.73305755853652954),
# ('rumination', 0.71992391347885132),
# ('distress', 0.7141801118850708),
# ('aggressive', 0.7049030065536499),
# ('craving', 0.70202392339706421),
# ('trait', 0.69775849580764771)]
It’s easy to see that corpus context is important - here is finding similar terms for the “reddit” corpus:
model = load_models(base_dir)["reddit"]
model.most_similar("anxiety")
# [('crippling', 0.64760375022888184),
# ('agoraphobia', 0.63730186223983765),
# ('generalized', 0.61023455858230591),
# ('gad', 0.59278655052185059),
# ('hypervigilance', 0.57659250497817993),
# ('bouts', 0.56644737720489502),
# ('depression', 0.55617612600326538),
# ('ibs', 0.54766887426376343),
# ('irritability', 0.53977066278457642),
# ('ocd', 0.51580017805099487)]
Here are examples of performing addition and subtraction with vectors:
model.most_similar(positive=['anxiety',"food"])
# [('ibs', 0.50205761194229126),
# ('undereating', 0.50146859884262085),
# ('boredom', 0.49470821022987366),
# ('overeating', 0.48451068997383118),
# ('foods', 0.47561675310134888),
# ('cravings', 0.47019645571708679),
# ('appetite', 0.46869537234306335),
# ('bingeing', 0.45969703793525696),
# ('binges', 0.44506731629371643),
# ('crippling', 0.4397256076335907)]
model.most_similar(positive=['bipolar'], negative=['manic'])
# [('nos', 0.36669495701789856),
# ('adhd', 0.36485755443572998),
# ('autism', 0.36115738749504089),
# ('spd', 0.34954413771629333),
# ('cptsd', 0.34814098477363586),
# ('asperger', 0.34269329905509949), ('schizotypal', 0.34181860089302063),
('pi', 0.33561226725578308), ('qualified', 0.33355745673179626),
('diagnoses', 0.32854354381561279)]
model.similarity("anxiety","depression")
#0.67751728687122414
model.doesnt_match(["child","autism","depression","rett","cdd","young"])
#'depression'
And to get the raw vector for a word:
model["depression"]
Extracting term similarities
To extract a pairwise similarity matrix, you can use the function extract_similarity_matrix. These are the data driven relationships between terms that the Wordfish infrastructure provides:
# Extract a pairwise similarity matrix
wordfish_sims = extract_similarity_matrix(models["neurosynth"])
Classification
Finally, here is an example of predicting neurosynth abstract labels using the pubmed neurosynth corpus. We first want to load the model and meta data for neurosynth, meaning labels for each text:
model = load_models(base_dir,"neurosynth")["neurosynth"]
meta = get_meta(base_dir)["neurosynth"]
We can then use the featurize_to_corpus method to get labels and vectors from the model, and the build_svm function to build a simple, cross validated classified to predict the labels from the vectors:
vectors,labels = featurize_to_corpus(model,meta)
classifiers = build_svm(vectors=vectors,labels=labels,kernel="linear")
The way this works is to take a new post from reddit with an unknown label, use the Word2vec word embeddings vector as a lookup, and generating a vector for the new post based on taking the mean vector of word embeddings. It’s a simple approach, could be improved upon, but it seemed to work reasonably well.
Classification of Disorder Using Reddit
A surprisingly untapped resource are Reddit boards, a forum with different “boards” indicating a place to write about topics of interest. It has largely gone unnoticed that individuals use Reddit to seek social support for advice, for example, the Depression board is predominantly filled with posts from individuals with Depression writing about their experiences, and the Computer Science board might be predominantly questions or interesting facts about computers or being a computer scientist. From the mindset of a research scientist who might be interested in Reddit as a source of language, a Reddit board can be thought of as a context. Individuals who post to the board, whether having an “official” status related to the board, are expressing language in context of the topic. Thus, it makes sense that we can “learn” a particular language context that is relevant to the board, and possibly use the understanding of this context to identify it in other text. Thus, I built 36 word embedding models across 36 Reddit boards, each representing the language context of the board, or specifically, the relationships between the words. I used these models to look at context of words across different boards. I also build one master “reddit” model, and used this model in the classification framework discussed previously.
For the classification framework, it was done for two applications - predicting reddit boards from reddit posts, and doing the same, but using the neurosynth corpus as the Word2Vec model (the idea being that papers about cognitive neuroscience and mental illness might produce word vectors that are more relevant for reddit boards about mental illness groups). For both of these, the high level idea is that we want to predict a board (grouping) based on a model built from all of reddit (or some other corpus). The corpus used to derive the word vectors gives us the context - meaning the relationships between terms (and this is done across all boards with no knowledge of classes or board types), and then we can take each entry and calculate an average vector for it based on averaging the vectors of word embeddings that are present in the sentence. Specifically we:
1) generate word embeddings model (M) for entire reddit corpus (resulting vocabulary is size N=8842) <br>
2) For each reddit post (having a board label like "anxiety":<br>
- generate a vector that is an average of word embeddings in M<br>
Then for each pairwise board (for example, “anxiety” and “atheist”
1) subset the data to all posts for “anxiety” and “atheist”
2) randomly hold out 20% for testing, rest 80% for training
3) build an SVM to distinguish the two classes, for each of rbf, linear, and poly kernel
4) save accuracy metrics
Results
How did we do?
Can we classify reddit posts?
The full result has accuracies that are mixed. What we see is that some boards can be well distinguished, and some not. When we extend to use the neurosytnh database to build the model, we don’t do as well, likely because the corpus is much smaller, and we remember from the paper that larger corpus tends to do better.
Can we classify neurosynth labels?
A neurosynth abstract comes with a set of labels for terms that are (big hand waving motions) “enriched.” Thus,given labels for a paragraph of text (corresponding to the neurosynth term) I should be able to build a classifier that can predict the term. The procedure is the same as above: an abstract is represented as its mean of all the word embeddings represented. The results are also pretty good for a first go, but I bet we could do batter with a multi-class model or approach.
Do different corpus provide different context (and thus term relationships?)
This portion of the analysis used the Word2Vec models generated for specific reddit boards. Before I delved into classification, I had just wanted to generate matrices that show relationships between words, based on a corpus of interest. I did this for NeuroSynth, as well as for a large sample (N=121862) reddit posts across 34 boards, including disorders and random words like “politics” and “science.” While there was interesting signal in the different relationship matrices, really the most interesting thing we might look at is how a term’s relationships varies on the context. Matter of fact, I would say context is extremely important to think about. For example, someone talking about “relationships” in the context of “anxiety” is different than someone talking about “relationships” in the context of “sex” (or not). I didn’t upload these all to github (I have over 3000, one for each neurosynth term), but it’s interesting to see how a particular term changes across contexts.
Each matrix (pdf file) in the folder above is one term from neurosynth. What the matrix for a single term shows is different contexts (rows) and the relationship to all other neurosynth terms (columns). Each cell value shows the word embedding of the global term (in the context specified) against the column term. The cool thing for most of these is that we see general global patterns, meaning that the context is similar, but then there are slight differences. I think this is hugely cool and interesting and could be used to derive behavioral phenotypes. If you would like to collaborate on something to do this, please feel free to send me an email, tweet, Harry Potter owl, etc.
Conclusions
Wordfish provides standard data structures and an easy way to extract terms, corpus, and perform a classification analysis, or extract similarity matrices for terms. It’s missing a strong application. We don’t have anything suitable in cognitive neuroscience, at least off the bat, and if you might have ideas, I’d love to chat. It’s very easy to write a plugin to extend the infrastructure to another terminology or corpus. We can write one of those silly paper things. Or just have fun, which is much better. The application to deploy Wordfish plugins, and the plugins themselves are open source, meaning that they can be collaboratively developed by users. That means you! Please contribute!.
Limitations
The limitations have to do with the fact that this is not a finished application. Much fine tuning could be done toward a specific goal, or to answer a specific question. I usually develop things with my own needs in mind, and add functionality as I go and it makes sense.
Database
For my application, it wasn’t a crazy idea to store each corpus entry as a text file, and I had only a few thousand terms. Thus, I was content using flat text files to store data structures. I had plans for integration of “real” databases, but the need never arose. This would not be ideal for much larger corpus, for which using a database would be optimal. Given the need for a larger corpus, I would add this functionality to the application, if needed.
Deployment Options
Right now the only option is to generate a folder and install on a cluster, and this is not ideal. Better would be options to deploy to a local or cloud-hosted virtual machine, or even a Docker image. This is another future option.
Data
It would eventually be desired to relate analyses to external data, such as brain imaging data. For example, NeuroVault is a database of whole-brain statistical maps with annotations for terms from the cognitive atlas, and we may want to bring in maps from NeuroVault at some point. Toward this aim a separate wordfish-data repo has been added. Nothing has been developed here yet, but it’s in the queue.
And this concludes my first un-paper paper. It took an afternoon to write, and it feels fantastic. Go fish!
Suggested Citation:
Sochat, Vanessa. "Wordfish: tool for standard corpus and terminology extraction." @vsoch (blog), 19 Feb 2016, https://vsoch.github.io/2016/2016-wordfish/ (accessed 03 Jan 26).