An important aspect of using containers in science is reproducibility, but it’s possibly not enough. It’s a good start to package your software and other dependencies in a container, but given that we are also interested in answering a scientific question, we need to understand why particular changes to a container might lead to changes in our result. In early development and thinking about Singularity Hub and container metrics this work was started by discussing various levels of reproducibility. At the time, I had ideas for metrics, but was lacking in containers. It’s now been a few years and we have happily been building up a database of over 1,000 containers, and now what we need to do is clear. These are the three buckets of things that I want to talk about today:
- Tools for visualization, summary, or general introspection to better understand research software.
- Community data resources to expose the raw data directly to you
- Applied starter examples and templates to inspire, or just use as is.
No, I’m not saying I want to do all this myself! This isn’t the effort for a single dinosaur, it must come from the power of our community, and the community must be empowered by the software engineers that create tools that help drive it (cue Circle of Life from the Lion King).
Container Diff, No it’s not “C-Diff”
Tool to produce programmatically accessible introspection of containers
Our story starts around the same time as the Singularity Hub paper. Google came out
with a tool called Container Diff, and
I was crazy excited because I wanted others to see how important introspection and understanding of these black boxes truly is. The tool could
work with various kinds of containers, but not the technology (Singularity)
that High Performance Computing (HPC) Centers and general researchers
can use on their shared resources. To help with this, and to leverage this awesome tool, I want to present a Singularity wrapper to container-diff, a script called
analyze-singularity.sh that will take a Singularity container and send it through the same
process. In fact, we export the container as a .tar file and then use the same container-diff tool so the output is equivalent. From this I was very
easily able to do things like use an information coefficient to compare differences between Ubuntu container versions.
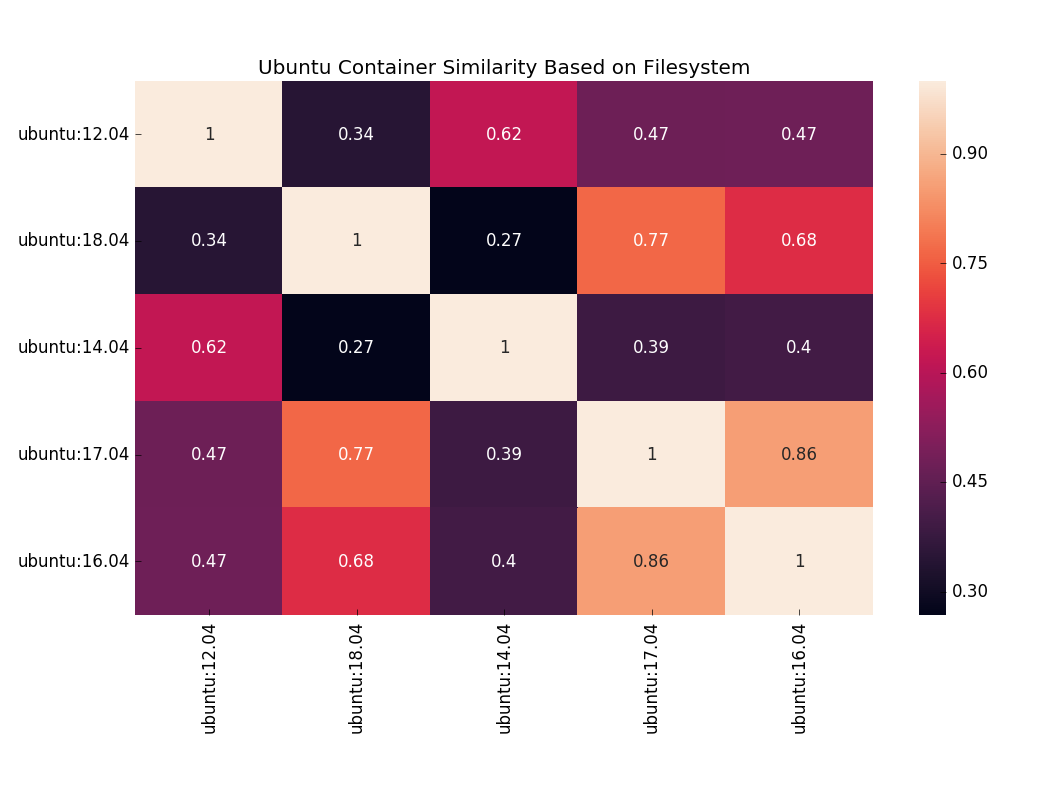
Plot courtesy of seaborn!
Want to use a different metric to compare files, or do something with packages? Or package versions? or files sizes? The data is all there to do it! Or want to generate data for your own custom set of containers? Well you can do that too, because I provided scripts and examples in that folder. But then I realized that it wasn’t really good enough to give you just scripts and examples. I wanted to:
- Make a container that takes your container choices, for Docker and/or Singularity
- Produces data to generate the visualizations (keep them keep if you want!)
- Create a simple web interface to show a similarity matrix, along with interactive filesystem views to inspect changes.
And of course I needed to put the entire thing in a container so you wouldn’t need to install things, other than maybe Docker. Yeah so, this is a new thing too. But this was actually a few days ago, and I got distracted again! While there are some things left to do, the gut of the project is there and I want to share with you to start this conversation about understanding containers. The delay in this post (note the original date was May 1st) is because I realized that instead of requiring you to derive data for containers, I should provide it as a resource. Enter the Container API!
The Container Differences API
Community data resources to expose the raw data directly to you
Playing with data is fun. Given a nicely formatted dataset, it’s a fun evening activity to load into your favorite software of choice and play around with. The statement above calls for the following needed features:
A Community Resource
We need a community data resource to serve enough metadata about our containers for you to do analyses. While Singularity Hub serves basic metrics and files, in the long term I don’t want the build service to have any extra burden in serving large metadata files. Singularity Hub must be optimized to serve the containers themselves for our academic community. This calls for some separate resource to serve the metadata.
Programmatic Accessibility
I want this resource to be available, in different formats, to every researcher large and small. It goes without saying, then, that the resource must be programmatically accessible, and not just “follow this cryptic set of instructions to get it in this one way” but rather “we’ve thought about the ways you already use to get information, and you can do that here too.” What does this look like, for a lot of you? You are going to be doing your analysis in likely one of a handful of popular “scientific software” suites, such as Python, R, Julia, Matlab, or perhaps using old school curl or wget on the command line to retrieve the resource.
Github for Accessibility and Version Control
Something being programmatically accessible doesn’t mean everyone can get at it. For example, I barely knew what an API was when I started graduate school, let alone used them. I did, however, know about Github cloning. Can our data be dually served on Github (with version control) and from APIs? It can if it’s the same data! If we have a repository that serves a static json API that you can also clone, we kill two birds with one stone. We are able to support tagging of versions and discussion, which is just more awesomeness. If I want to make a copy, I can just clone it. I can write other tools around it using Github’s APIs.
To make this possible I’ve started the Container Static Community API. You can literally go to one of the endpoints (for example, for files) and instantly get access to the entire filesystem contents and sizes for public images in Singularity Hub. Do you want a list of packages and versions? Yeah, we have that too, along with an inspect (that has the recipe and definition file). Is this a sophisticated, serverless technology with crazy expensive hosting and a flashy web interface? No, it’s the Github that we know and love! So what can you do with this? Here are some questions that are of great interest to me, and I bet you would enjoy working on. If you want a collaborator, you know how to find me :)
1. We have not tested robust ways to compare containers. You might test comparison metrics over a set of container types and outcomes, and help to uncover what kinds of measurements are important toward some goal.
2. Do you want to optimize building? Running? Or just learn about container recipe practices in general? You can get started.
3. What kind of software and environments are we putting in containers? Can we track trends? (Tensorflow I’m looking at you!)
4. How do people document (or don’t) their containers?
5. How do containers relate to one another? Do you see common bases, or does everyone reinvent the wheel?
6. How do we curate containers? What is it based on?
There are truly endless interesting questions that we might ask, and they all start with data. They are incredibly challenging because the data we are working with is living and constantly changing, and we never stop to ask how to computer we’re on today is comparable to the “same” flavor of operating system 15 years ago. I’ve been particularly interested in how operating systems change over time, and how code is generally used in research, and encourage you to poke me if you want to work on a fun project. I also want feedback from you about this API! Do you want a client to interact with it? I am trying to develop “infrastructure” that mirrors practices that researchers are familiar with. Please post an issue with your thoughts!
Getting Started Examples
To demonstrate using the analyze-singularity.sh with Google’s container-diff to develop a new kind of interactive visualization, I created
this example project, and it produces data:
├── packages.json
├── packages.tsv
├── ubuntu:12.04-files.json
├── ubuntu:12.04-packages.json
├── ubuntu:14.04-files.json
├── ubuntu:14.04-packages.json
├── ubuntu:16.04-files.json
├── ubuntu:16.04-packages.json
├── ubuntu:17.04-files.json
├── ubuntu:17.04-packages.json
├── ubuntu:18.04-files.json
└── ubuntu:18.04-packages.json
and a nice web interface to view it, here is a…
DEMO
I literally spit this out in under 48 hours, so please excuse the less than perfect user interface. I’m not a designer, let’s leave it at that, heh. What you will see is a tab separated file, with an information coefficient to compare some set of containers you’ve chosen:
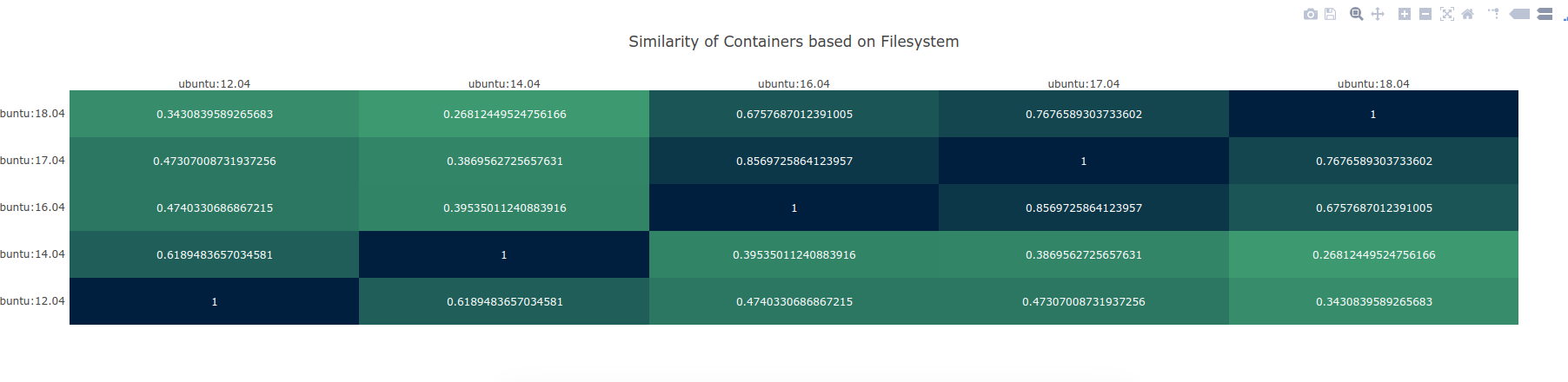
Plot courtesy of PlotlyJS
and then you can click on a comparison in a list at the bottom to view the additions / subtractions between any particular two!
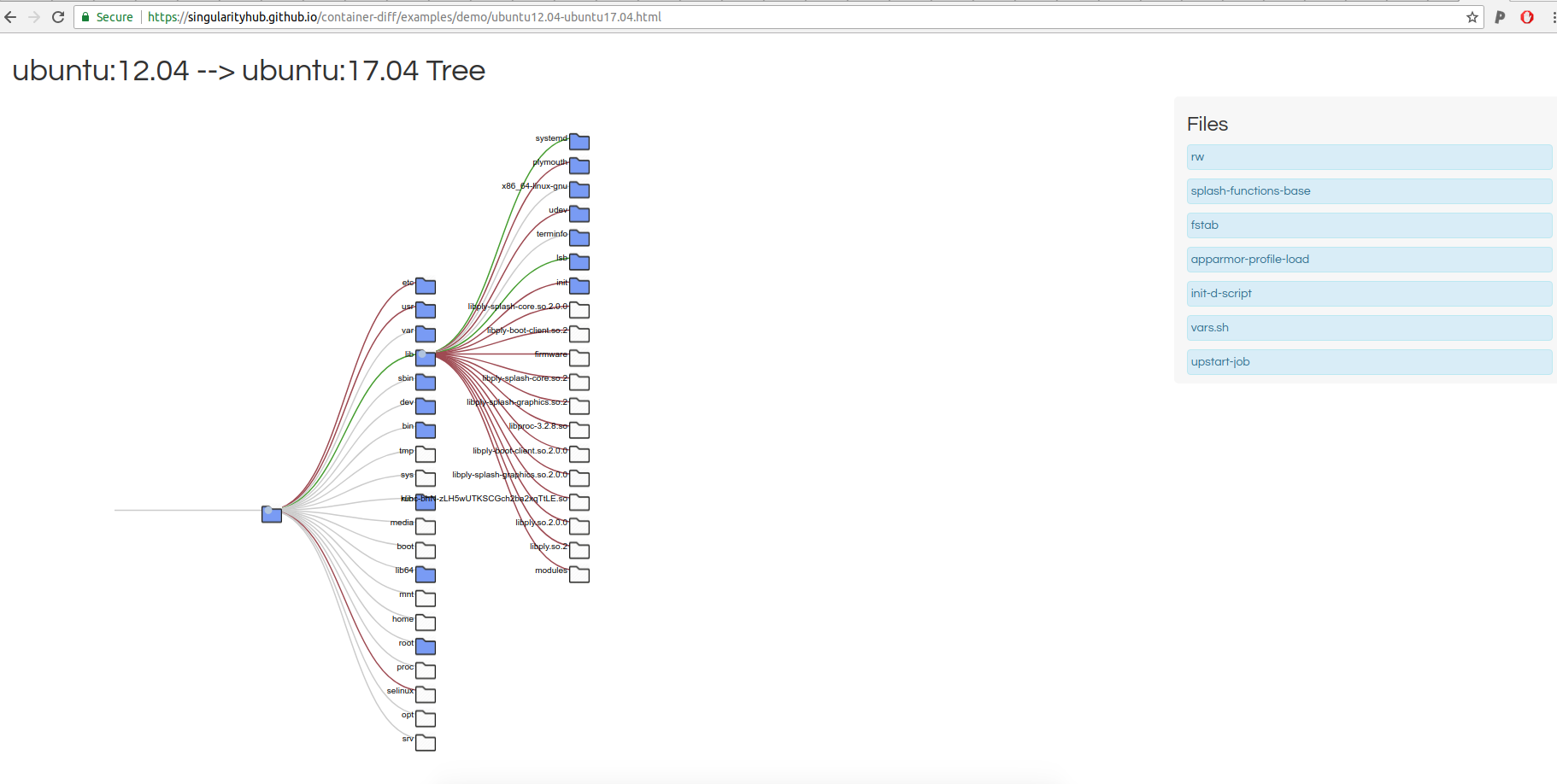
That entire thing would be produced with a command like this:
docker run -v /tmp/web:/data -p 8888:8888 -it vanessa/container-diff ubuntu:12.04 ubuntu:14.04 ubuntu:16.04 ubuntu:18.04
1. Staring extraction for 4 containers.
/data/ubuntu:12.04-files.json
/data/ubuntu:14.04-files.json
/data/ubuntu:16.04-files.json
/data/ubuntu:18.04-files.json
2. Calculating comparisons
Open browser to http://0.0.0.0:8888
I added the red and green at the last minute to show branches of added and removed content. And by the way, I’m already terribly critical of all of this, and have a list of issues on the queue to fix up. Small details like the size and spacing of the files list, the styling of the entire thing, and the ability to visualize and compare size are needed things. It’s also not optimized, between doing some very simple caching of results and not re-running if they already exist! For now, this serves as a simple example of a tool that I could make using the lower level comparison tools. These functions are served by Singularity Python, which means that you could swap out any interface or metric for your own work. If you can’t tell, I’m so eager and excited to share this with you. Cool!
Container Metrics and Operating System Science
Let’s get into the meat of this discussion. Singularity Python started as a “collect all” repository that I dumped various Python utils for working with Singularity images. I know Python isn’t flashy like some other new languages, but it’s near and dear to my heart, and I love using it. This little repository quickly turned into a number of small tools and clients, along with an image manager, primarily in response to user requests and needs and my own desires to manage and organize things. With Singularity Hub it took on the burden of more functions for building, comparison of containers and visualization, and then one day a user came to me and said “this is too much.” Although I knew it would be an insurmountable amount of work, I knew that the repository needed to be cleaned up. The tools needed to be separated, because separation would be essential to having the smallest number of dependencies for each. Isolated development was also important to not put any burden on any one particular tool that is more needed than another. For example, one package for calculating a date time offset was making users of Python 2 want to throw their pancakes at the sky with frustration. I wanted to develop at the same pace as the user need, and I was slowing myself down. If one tool has a slower release cycle why should the others that have bugs ready to fix wait on that? There are so many good and logical reasons for isolation and simplification. So that was the plan.
Now we have that separation! But where does that leave Singularity Python? It’s right back to where it started, actually, and in this direction it of container metrics and analysis it will continue to be developed! This kind of research is what I call operating system science and I want to study it. It’s what drove me to mine Pubmed Central for Github links as a graduate student (sorry to my lab for taking up all of the space on Sherlock for my “database”) and to now spend inordinate amounts of time thinking about how to see things that have much beauty hiding behind a command line.
I am hopeful that with tools, datasets, and fun spirit we can work on these things together, and better understand computers, containers, and generally software as living, changing entities.
I haven’t shown this to many, but back in summer 2016 (before I had graduated). I had vision for building tools and infrastructure for reproducible science. This is one of the many visions that lives somewhere on my frontal lobes that was, for this afternoon, splot out onto a whiteboard in a massive brain dump that smelled like dry erase markers:
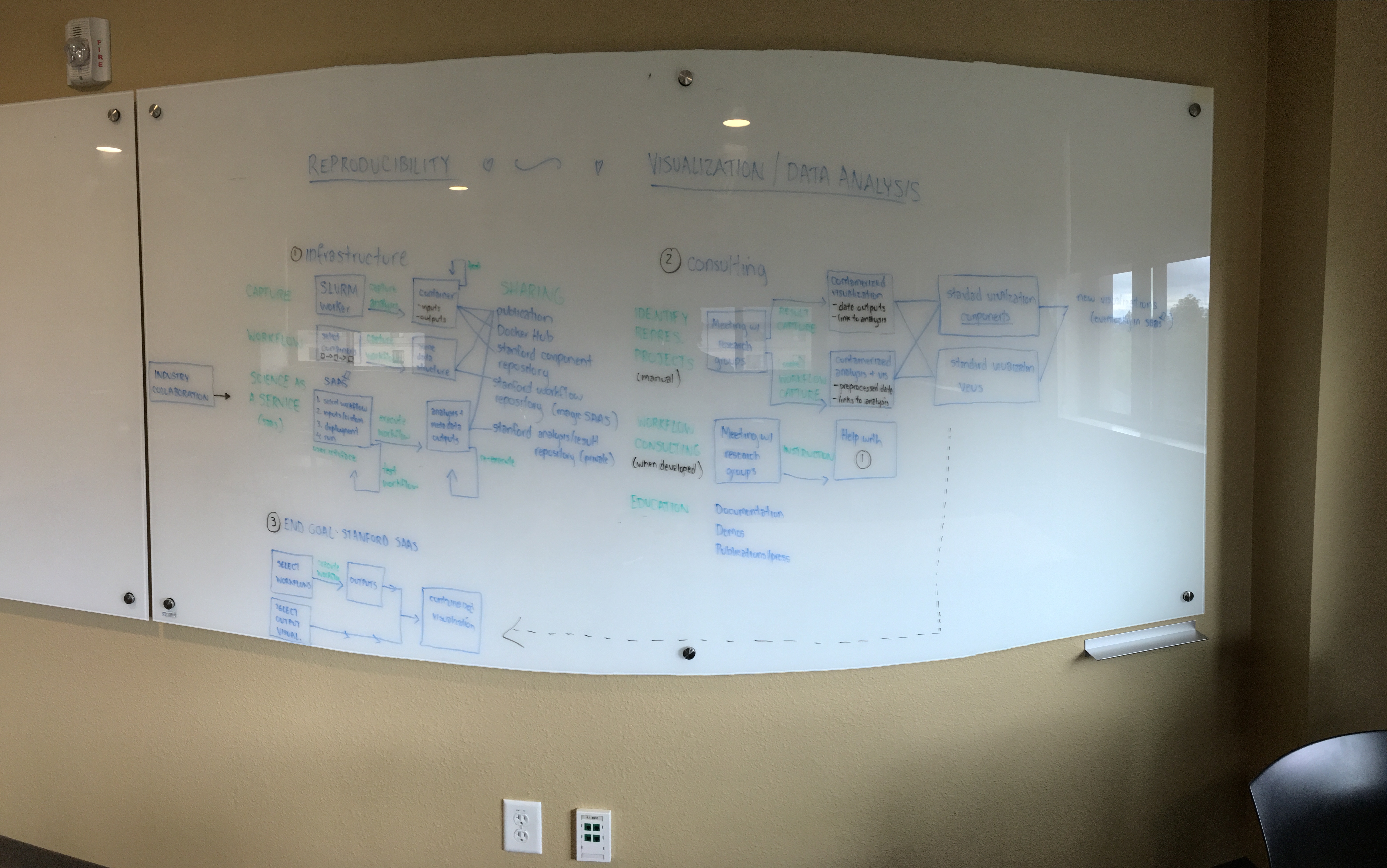
Can we talk about that board for a second? Does anyone understand this board design? its too shiny and “3D” to actually be able to read anything clearly on.
What I didn’t realize at the time is that the world wasn’t ready for the more advanced part of that vision (and in fact we still don’t have these “click button and run” and “science as a service” for most academic groups) but guess what? I see the very tangible, and clear steps for getting us there, and it’s going to happen sooner rather than later. Two years ago we needed to start small and just keep this larger vision in mind. Across domains of science we needed to convince other researchers that sharing data was a good thing to do, and having transparency in our work was the right way to do science, period. We started with just having a continer technology. or even more than one, and then a registry, and added on version control and testing to that. Now we have immense awareness, and this beautiful collective energy to work on these problems together. At the time I also wanted to work hard to have better collaborations between industry groups like Google and Stanford, and I’m seeing this happen on a regular basis now, whereas before it was a challenge. We are also getting away from that mindset of “cloud vs. HPC” and “my cluster is bigger than your cluster!” We all know it’s not the size of the cluster that matters… okay bad joke :)
All kidding aside, we have learned and done so much in just a short two years! It’s so wicked awesome. Let’s keep doing this, Ok? If you are a researcher and something is hard, and you need a tool, or Stanford is “missing this one thing” or “could we just…” I hope that you continue to reach out to me. It is my job to anticipate your needs and build tools that there might not be NIH funding for, but will improve your daily workflow by ten fold. If something isn’t good, then be vocal about it. If you need help, then you should ask for it, because we are here for you. Please give feedback to any and all that will listen about the importance of research software engineers! Supporting funding for research software engineers == better tools == better science. Do you want to know more about my group? We are here for you! From office hours to custom software to running your thing at scale, we are here to help! I’m terrible at so many things HPC, but guess what? My group, and our fearless leader, they blow me away on a daily basis. I sit here and cycle through Python, containers, and some other basic tooling, and am inspired. And guess what? I’m proud to announce our newest Research Computer member! Welcome to the newest Robot to join the “Robot Roll Call” :):

In Summary!
- The Container API is going to be a continually growing resource for you to do your own work to study containers.
- If you want to quickly compare or extra metadata for any set of Docker/Singularity/other containers, do it.
- Let's think together about container metrics, and continued development of tooling!
To keep everything tidy, Container Tools has a simple portal to take you to different documentation and repositories, and you can of course follow on Github or Twitter.
Party on, friends! Time for some sleep for this programming dinosaur.
Suggested Citation:
Sochat, Vanessa. "Container Differences." @vsoch (blog), 03 May 2018, https://vsoch.github.io/2018/container-diff/ (accessed 03 Jan 26).