Before we go into details, here is what I am going to talk about in this post:
- Container-diff is HPC Friendly. It affords doing a scaled analysis of Dockerfiles.
- ImageDefinition is my proposal to schema.org to describe a Dockerfile.
- I made Github Actions to extract metadata for your Dockerfiles and Datasets in Github repositories.
Let’s get started!
container-diff is HPC Friendly
When I first wanted to use Google’s container-diff at scale, I realized that it wasn’t suited for a cluster environment. Why? Because
1. I couldn’t control the cache, and it would default to my home that would subsequently fill
up almost immediately.
2. There wasn’t an
option to save to file, and I couldn’t come up with a (reasonably simple) solution to pipe the output
from a node. Sure, I could come up with a hack, but it wasn’t a long term solution.
To solve these problems, despite being super noobish with GoLang, I did two pull requests (now merged into master, 1 and 2) to add the ability to customize the cache and save output to a file. In a nutshell, I added command line options for an “–output” and “–cache-dir” and a few tests. The release with these features hasn’t yet been done yet at the time of writing of this post, so if you are interested in testing you would need to build from the master branch. I also want to say thank you to @nkubala because contributing was really fun. This is the best way to run open source software.
Analysis of Dockerfiles
Let’s cut to the chase. I filtered down the original 120K to 60K that had Python (pip) packages, and then did a small visualization of a subset that would be reasonable to work with on my local machine. In a nutshell, there is beautiful structure:
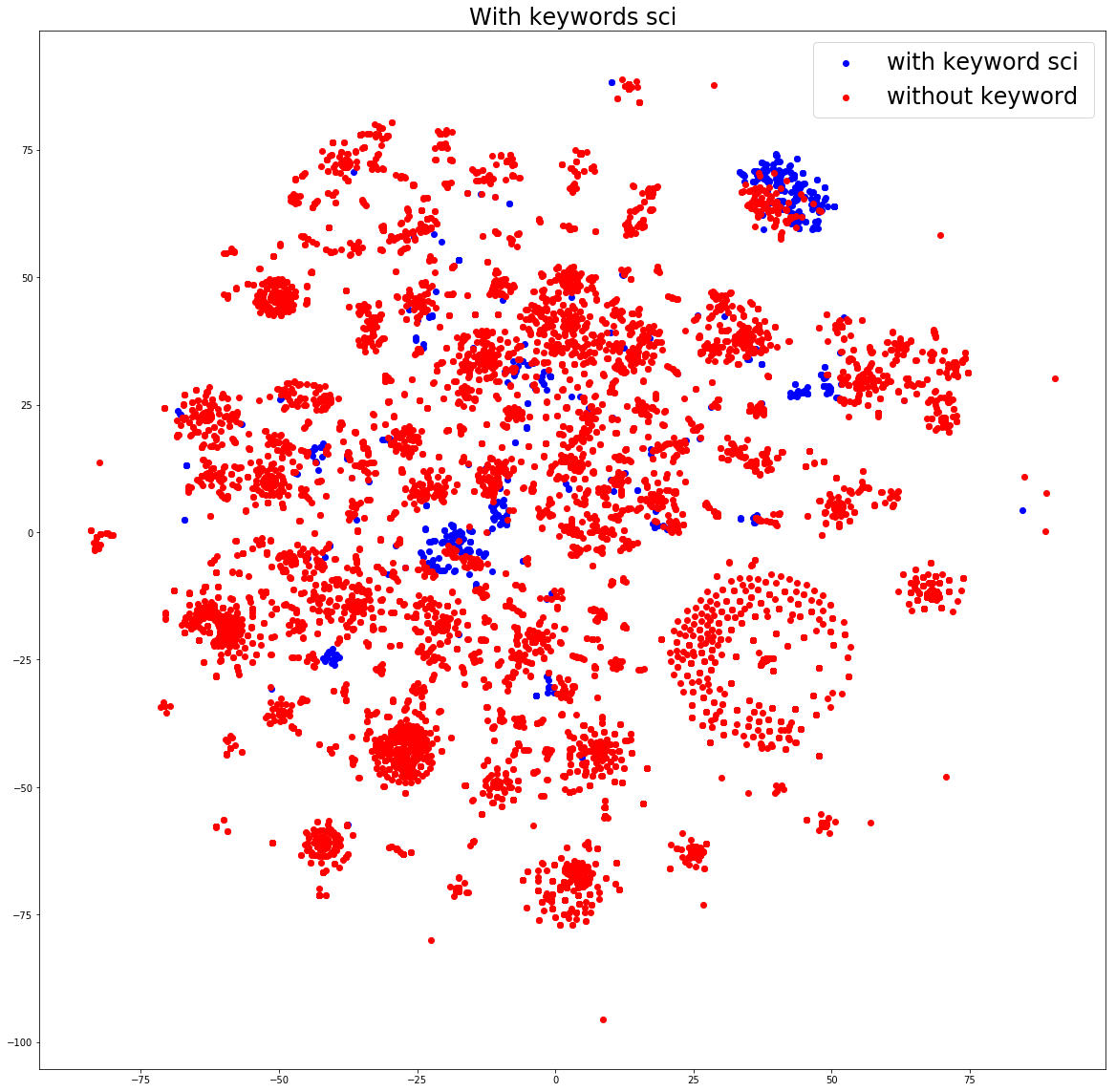
If you want to see the entire analysis, the notebook is available. The take away message that I want you to have is that containers are the new unit that we operate in. We don’t understand their guts. With tools like container-diff, and with data structures like an “ImageDefinition” (hopefully soon defined by schema.org), we can have all this metadata splot out statically in HTML for search giants like Google to parse. We might actually be able to parse and organize this massive landscape. How do you do that? Let me show you how to start.
How do we label a Dockerfile?
For each Dockerfile, I used schemaorg Python to define an extractor, and used the extractor to generate an html page (with embedded metadata). If you remember the early work thinking about a container definition and then doing an actual extraction this was leading up to the extraction done here - we finally labeled each Dockerfile as an “ImageDefinition”. The extractors I’m talking about are provided via containers served by the openschemas/extractors Github repository.
The ImageDefinition
This jumps to our next question. We needed an entity in schema.org to define a build specification / container recipe or definition like a Dockerfile. This is the “ImageDefinition”.
What is an ImageDefinition?
An ImageDefinition is my favorite proposed term for what we might call a container recipe, the “Dockerfile.” It is essentially a SoftwareSourceCode that has some extra fields to describe containers. I’ve been trying to engage various communities (Schemaorg and OCI) for a few months to take interest (imagine if Google Search could define and index these data structures! That alone would be huge incentive for others to add the metadata to their Github repos and other webby places).
To make it easy for you to extract ImageDefinitions and Datasets for your software and repositories, I’ve created a repository of extractors. Each subfolder there will extract a schema.org entity of a particular type, and create a static html file (or a json structure) to describe your dataset or Dockerfile. Here is a random example from the GIthub pages for that repository for a container Dockerfile that provides both Apt and Pip packages, extracted thanks to Container-diff, and parsed into that metadata thanks to schamaorg Python.
What is an Extractor?
An extractor is just a set of Python scripts that use schemaorg python to generate a datastructure (json or html with json embedded) to describe an entity described in schema.org like a Dataset or Software source code. You can use or modify the extractors scripts directly for your purposes, or for Dataset or ImageDefinition, follow the instructions in the repository READMEs to run a container locally:
$ docker run -e DATASET_THUMBNAIL=https://vsoch.github.io/datasets/assets/img/avocado.png \
-e DATASET_ABOUT="This Dockerfile was created by the avocado dinosaur." \
-e GITHUB_REPOSITORY="openschemas/dockerfiles" \
-e DATASET_DESCRIPTION="ubuntu with golang and extra python modules installed." \
-e DATASET_KWARGS="{'encoding' : 'utf-8', 'author' : 'Dinosaur'}" \
-it openschemas/extractors:Dataset extract --name "Dinosaur Dataset" --contact vsoch --version "1.0.0"
These containers on Docker Hub are what drive the Github Actions to do the same, discussed next.
Github Actions
But I wanted to make it easier. How about instead of tweaking Python and bash code, you had a way to just generate the metadata for your Github repos? This can be helped with Github actions!
How do I set up Github Actions?
It comes down to adding a file in your repository that looks like .github/main.workflow that then
gets run via Github’s internal Continuous Integreation service called Actions.
What goes in the main workflow?
Here are recipes and examples to show you each of the demos for ImageDefinition and Dataset, respectively.
Extract a Dataset
zenodo-ml describes the vsoch/zenodo-ml repository, with a bunch of Github code snippets from Zenodo records and their metadata. This is a Dataset and you can see it’s valid based on the Google Metadata Testing tool:
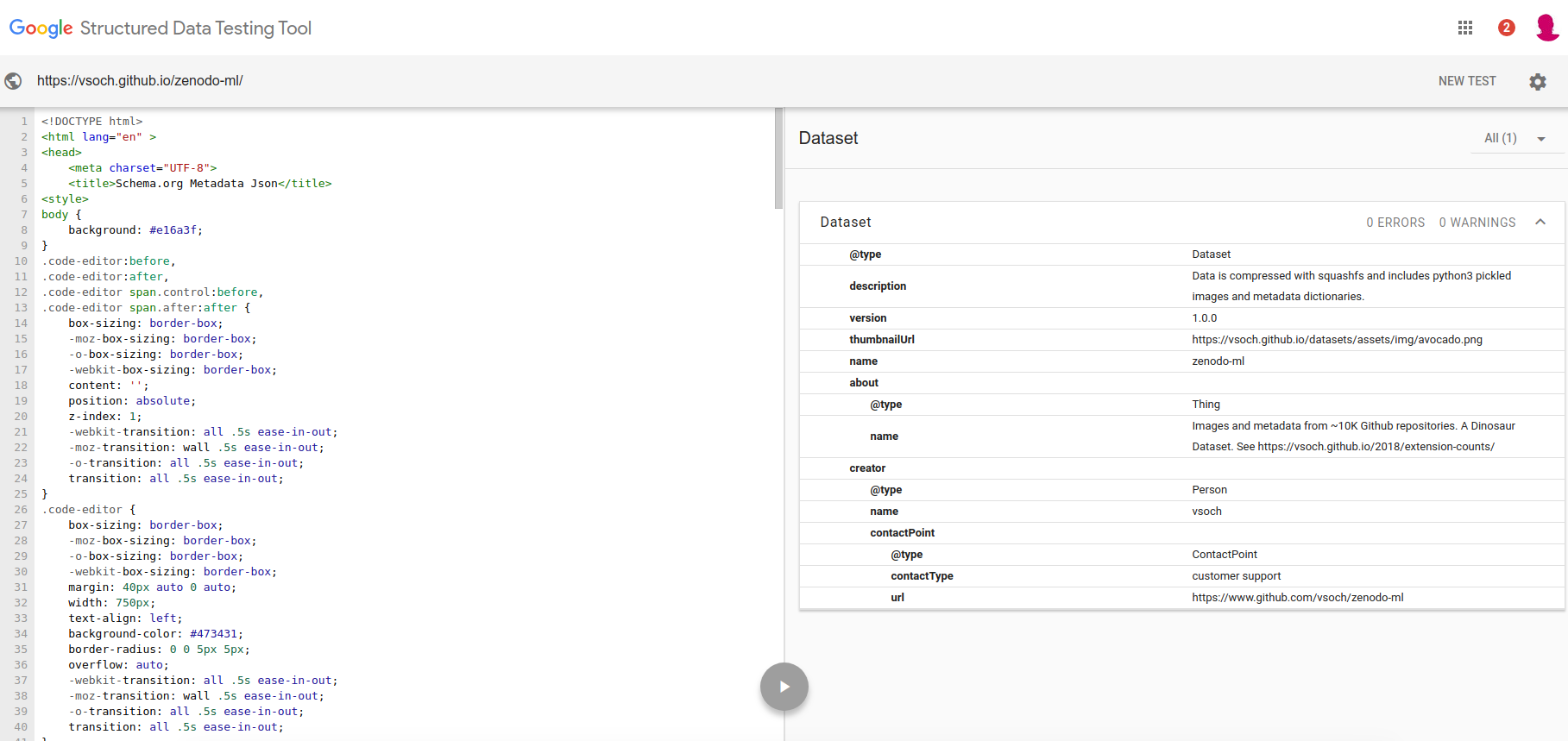
Here is a snapshot of the site that is validated. You can see the metadata there, and it’s also provided in a script tag for the robots to find.
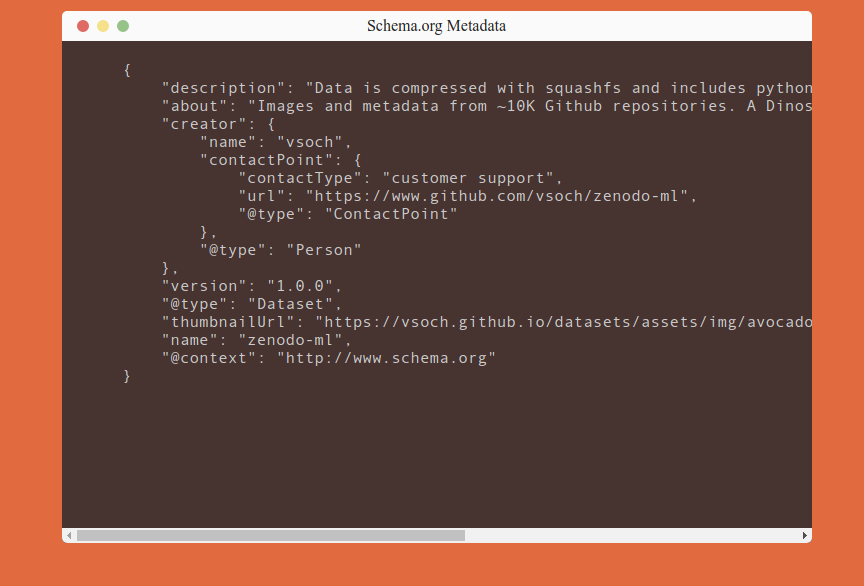
The main workflow looks like this:
workflow "Deploy Dataset Schema" {
on = "push"
resolves = ["Extract Dataset Schema"]
}
action "list" {
uses = "actions/bin/sh@master"
runs = "ls"
args = ["/github/workspace"]
}
action "Extract Dataset Schema" {
needs = ["list"]
uses = "docker://openschemas/extractors:Dataset"
secrets = ["GITHUB_TOKEN"]
env = {
DATASET_THUMBNAIL = "https://vsoch.github.io/datasets/assets/img/avocado.png"
DATASET_ABOUT = "Images and metadata from ~10K Github repositories. A Dinosaur Dataset."
DATASET_DESCRIPTION = "Data is compressed with squashfs, including images and metadata. "
}
args = ["extract", "--name", "zenodo-ml", "--contact", "@vsoch", "--version", "1.0.0", "--deploy"]
}
In the first block we define the name of the workflow, and we say that it happens on a “push.” In the second block, we just list the contents of the Github workspace. This is a sanity check to make sure that we have files there. In the third block, we do the extraction! You’ll notice that I am using the Docker container openschemas/extractors:Dataset and that several environment variables are used to define metadata, along with command line arguments under args. I’m not giving extensive details here because I’ve written up how to write this recipe in detail in the README associated with the action. The last step also deploys to Github pages, but only given that we are on master branch.
Extract an ImageDefinition
As an example of an ImageDefinition, I chose my vsoch/salad repository
that builds a silly container. You can see it’s ImageDefinition here.
It’s also generated via a Github Action. The main workflow looks like this:
workflow "Deploy ImageDefinition Schema" {
on = "push"
resolves = ["Extract ImageDefinition Schema"]
}
action "build" {
uses = "actions/docker/cli@master"
args = "build -t vanessa/salad ."
}
action "list" {
needs = ["build"]
uses = "actions/bin/sh@master"
runs = "ls"
args = ["/github/workspace"]
}
action "Extract ImageDefinition Schema" {
needs = ["build", "list"]
uses = "docker://openschemas/extractors:ImageDefinition"
secrets = ["GITHUB_TOKEN"]
env = {
IMAGE_THUMBNAIL = "https://vsoch.github.io/datasets/assets/img/avocado.png"
IMAGE_ABOUT = "Generate ascii art for a fork or spoon, along with a pun."
IMAGE_DESCRIPTION = "alpine base with GoLang and PUNS."
}
args = ["extract", "--name", "vanessa/salad", "--contact", "@vsoch", "--filename", "/github/workspace/Dockerfile", "--deploy"]
}
The only difference from the above (primarily) is that we build our vanessa/salad container first,
and then run the extraction.
Github Bugs
There are some bugs you should know about if setting this up! When you first deploy to Github pages, the permissions are wrong so that it will look like your site is rendering Github pages, but you will get a 404. The only fix is to (in your settings) switch to master branch, and then back to Github Pages. The same thing unfortunately happens on updates - you will notice the Github Action runs successfully, but then that the page isn’t updated. If you go back to settings you will see the issue:
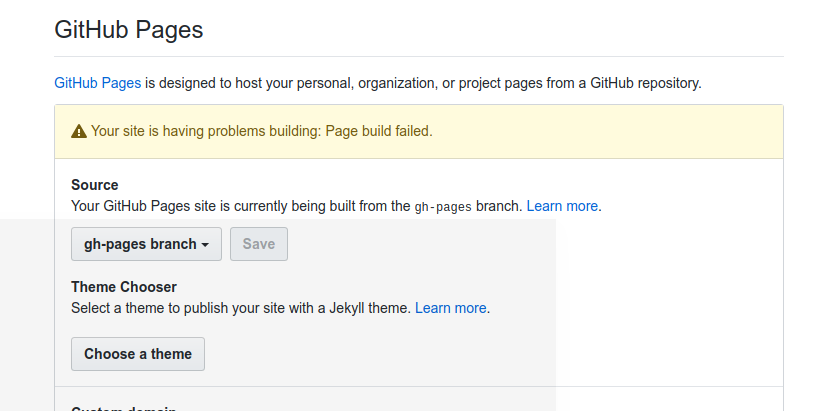
And again the fix is to switch back and forth from master back to Github pages.
Summary
I hope that I’ve started to create something that could be useful. Please open issues if you have questions or want to help to make the tools better. What can you do to actively start exposing the metadata for your Datasets and Containers? Here are some suggestions:
- Use the extractors locally or via Github actions.
- Do a better job than me to analyze all those Dockerfiles!
- Use container-diff to do your own analysis
- Analyze software via code in Github repositories with the zenodo-ml dataset.
Dinosaur out.
Suggested Citation:
Sochat, Vanessa. "The Ultimate Dockerfile Extraction." @vsoch (blog), 20 Dec 2018, https://vsoch.github.io/2018/image-definition/ (accessed 03 Jan 26).