In this post, I’m going to talk about the Scientific Filesystem Container Builder, a template for researchers to use to make it easy to distribute reproducible, tested research software. Yes, that means a workflow of:
build --> test --> deploy
The technologies that we use are Github for code and version control, CircleCI for testing, and Docker Hub for final container deployment. And guess what? You don’t need to do much more than fork and clone a Github repository, add your special software, and then connect to the services above. If you want to jump ahead, here is what we will review today:
Let’s go!
Background
Earlier this year I created the Scientific Filesystem (SCIF) as a means to make it easy to build containers for scientific applications. In a nutshell, SCIF let’s you install a single recipe into a container (any container technology) and then expose multiple entrypoints. An entrypoint, which is associated with app has it’s own context, including environment, runscript, tests, labels, and install routine. Given that a container has a scientific filesystem, the apps are always discoverable, inspectable, and usable for the user without needing to know a thing..
$ docker run vanessa/cowsay apps
animal
cowthink
train
message
list
fortune
md5sum
What makes them discoverable?
You can loop through that list, or use any of the entire environment namespace to interact with whatever application the user has decided to run, or just inspect the entire thing:
docker run vanessa/cowsay inspect
{
"animal": {
"appenv": [
"export PATH=/usr/games:$PATH"
],
"apptest": [
"export PATH=/usr/games:$PATH",
"export PATH=/usr/games:$PATH",
" exec cowsay -f vader-koala"
...
How does this happen?
It’s a bunch of work for the scientist to do, right? Not exactly. The scientist doesn’t need to know about how to write complicated entrypoint scripts for a container, how to set up testing, or how to format json and expose metadata. he or she can just fill in text blobs to define one or more of these sections:
%appinstall avocado
# Commands to install the software go here
%appfiles avocado
# You can add files from the host here that are needed for install, etc.
%apphelp avocado
# You can write a snippet of text to show to user for help.
%apprun avocado
# Commands executed when the app is run
%apptest avocado
# Commands to test your app, return non zero code if fail
%applabels avocado
MYAPP ISAWESOME
%appenv avocado
export NOMNOM=AVOCADO
And then you would put that in a recipe.scif (you can define as many apps as you
want, with as few or many of those sections as needed). This gets installed to a container (or host, shown later) and then interaction with the entire suite of applications is just via the single scif entrypoint, for example:
# List all apps installed
$ scif apps
$ scif version
# Commands
$ scif run avocado
$ scif exec avocado ls /scif/apps
$ scif test avocado
$ scif shell avocado
$ scif pyshell avocado
$ scif inspect avocado
There are a suite of commands for development, but I’ll skip over them for now. Let’s dive more into the rationale for the template we are going to describe today.
Building Containers and Testing is Hard
My first small project with SCIF was to build the same scientific workflow across 5 or 6 different container technologies. But I forgot about a few things.
- Building containers is hard
- Testing, and writing CircleCI or TravisCI configuration files is harder
- Creating a build --> test --> deploy setup is hard
It hit me like bird turd out of the sky that I could create a build –> test –> deploy template that handles all these hard things, and by way of having the scientific filesystem, the generation and testing would just be discovered.
Introducing the SCIF Container Builder!
The user can simply clone a repository template, write their own recipe, and then in a few clicks deploy a reproducible container with their software of choice! Inspired by some weekend project work where I finally figured out the CircleCI Version 2.0 Workflows setup I created the Scientific Filesystem Container Builder. It comes down to:
1. forking and cloning
2. Writing a recipe for your tool, and
3. connecting to Circle CI, and optionally defining a Docker Hub repo to deploy.
And that’s seriously it. If you want the superly verbose walkthrough to do this, see the builder documentation. For the rest of this post, I’ll briefly show you (pictures!) what is described there.
Template Overview
The idea of a repository as a template is not new. Having an example is the quickest way for someone to get comfortable with your technology, whether it’s a website theme, or a skeleton to build an API.
Github Templates to the Rescue!
In this template, I’m including these basic guts:
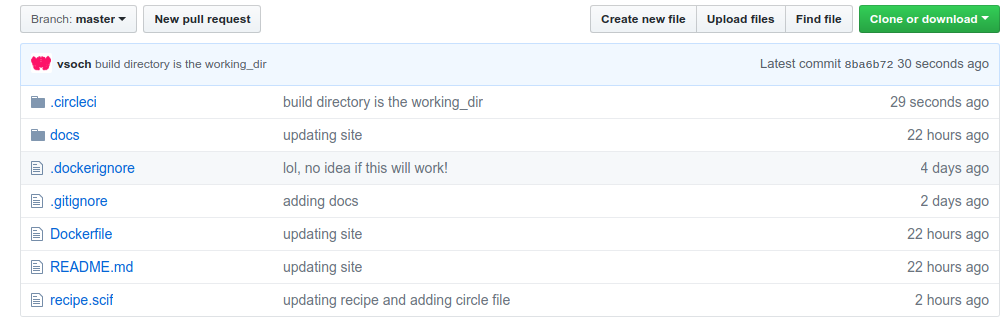
The recipe.scif is where the researcher writes the magic to install, describe, and test the research applications. Then in the Dockerfile it’s ready to go to install the recipe to a container. See the “hidden” .circleci folder? That has a really hairy config.yml file that defines multiple steps in a workflow that will build, test, and deploy. It looks like this:
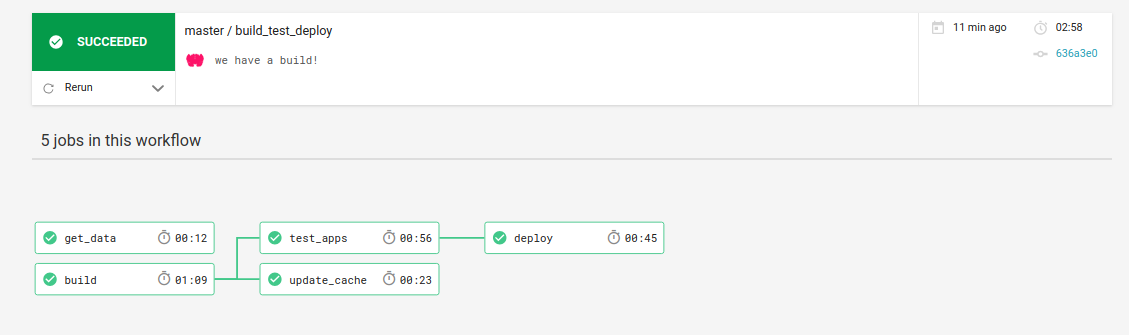
Don’t be afraid, you don’t need to edit this unless you want to add your own cool features.
Testing
Are you worried about testing? You just need to write some code snippets (or run a command) in
an %apptest <name> section, and that’s it! In the testing part of the workflow,
the apps are discovered in the container with scif apps and then looped through running
scif test app. This executes the code you’ve written! If the status code returned is 0 (meaning that you ran something and it didn’t error) the tests will pass. But guess what? SCIF doesn’t let you skimp! If you run the command to test and you didn’t write a test… YOU SHALL NOT PASS! Here is what happens when I tried to build without defining a test for the app “message”
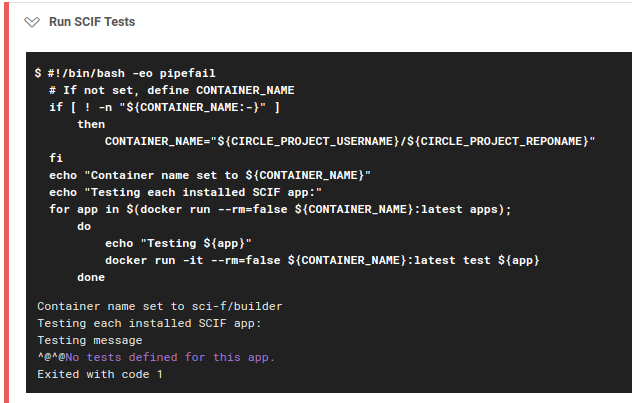
I’ve done this because the entire purpose of this setup is to test before deployment. Minimally, you can run the application entrypoint, or write some command that runs successfully.
%apptest animal
exec cowsay -f "hello-kitty"
My hope is that you will write an actual test.
Features
If you aren’t afraid of looking at the configuration files, you can customize and add your own cool features! This is a skeleton for a starter template, so I’m hopeful that you might (and share what you do!) Here are some examples.
Data Download
There is a (currently empty) section called get data where you might want to download and cache some external data source.
Metadata
There is another run section that will print the entire metadata dump for the container, here, and you might want to POST that somewhere, or otherwise use it.
Environment
Guess what, there is an entire namespace of environment variables available to use in the recipe, available during build and runtime of your application. This would be helpful to, for example, programmatically parse through metadata, identify the active app, or to generate a general application or function in the container that can use the environment variables to interact with the active (or sleeping) applications.
Deployment
The Dockerfile drives the build step that happens first, meaning that the recipe.scif
is built into the container, and the layers cached. Advanced users can add files, or change
the base image, or any other customizations (that aren’t associated with installing applications). The guts of Dockerfile looks like this (I’ve removed unnecessary notes):
FROM continuumio/miniconda3
# Install scif from pypi
RUN /opt/conda/bin/pip install scif==0.0.75
# Install the filesystem from the recipe
ADD *.scif /
RUN scif install /recipe.scif
# SciF Entrypoint
ENTRYPOINT ["scif"]
Base Image
I use continuumio because scif requires Python, and I like this base. You could change the base to be vanilla Python, or a smaller container all together. It’s overly verbose to basically install scif, add the recipe to the container, and install it.
Files
If you wanted to add files (for the recipes to use) you could do that like this:
ADD data/run.sh /
Which would mean that there is a data folder in the base of the repository, and
you are adding it to the root of the container. In the recipe.scif you could then
install it to an app like this:
%appfiles avocado
/run.sh
And it would be placed in the /scif/apps/avocado folder.
How can I add something to an app $PATH?
When a section is defined, a bin and lib are automatically generated for the app in question! The bin is added to the $PATH when the app is active, and lib to LID_LIBRARY_PATH. Thus, to make this script automatically found, just move it to the bin!
%appinstall avocado
mv run.sh bin/
chmod u+x run.sh bin/run.sh
Entrypoint
The scif entrypoint is the way to get access to all the commands like “run” “exec,” “inspect,” “shell,” etc. If you don’t specify it, it will show you these options! Since this is the entrypoint for the container, we can just run the container.
$ docker run vanessa/cowsay
Scientific Filesystem [v0.0.75]
usage: scif [-h] [--debug] [--quiet] [--writable]
{version,pyshell,shell,preview,help,install,inspect,run,test,apps,dump,exec}
...
scientific filesystem tools
optional arguments:
-h, --help show this help message and exit
--debug use verbose logging to debug.
--quiet suppress print output
--writable, -w for relevant commands, if writable SCIF is needed
actions:
actions for Scientific Filesystem
{version,pyshell,shell,preview,help,install,inspect,run,test,apps,dump,exec}
scif actions
version show software version
pyshell Interactive python shell to scientific filesystem
shell shell to interact with scientific filesystem
preview preview changes to a filesystem
help look at help for an app, if it exists.
install install a recipe on the filesystem
inspect inspect an attribute for a scif installation
run entrypoint to run a scientific filesystem
test entrypoint to test an app in a scientific filesystem
apps list apps installed
dump dump recipe
exec execute a command to a scientific filesystem
Deploy away, Merrill!
Currently, the last step to deploy requires definition of some environment variables in
your project. The user must define a DOCKER_USER and DOCKER_PASS in order for the final step (deployment to Docker Hub) to happen. If your Docker Hub repository that you’ve created is different from your Github username and password (likely) you also need to define a CONTAINER_NAME. The deploy will only happen on successful test after merge of master branch. The repository also needs to be created in the interface. When you finish, you should see these variables:
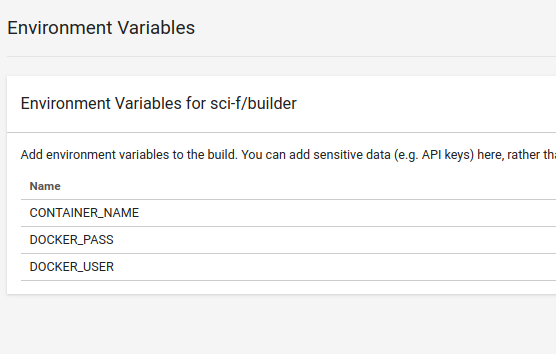
Take a look at the container built for this repository here.
Play with the Example
The example container is rather silly, you can see it here. Here is a quick show of it in action:
$ docker run vanessa/cowsay
Future Work
The amazing thing about this simple setup is that it affords so many different use cases! Here are just a few ideas.
Container Builders
Right now we are lacking in being able to label and then find our containers. Docker doesn’t expose it’s entire catalog endpoint, and even if it did, I’m not sure that a bunch of tags would be able to tell me the difference between the ten gazillion tensorflow images. Check this out:
Uhh… so which one is going to work for me? With a builder template like this, given that we have a simple metadata server somewhere, we would POST our metadata there on each automated deployment.
Workflow Modules
My original example with SCIF had a SCIF container wrapping a workflow, but really a good level of modularity is to imagine the entire container (with multiple possible entrypoints) as a block in the workflow. The reason for this is because we would want to distribute one reproducible container binary, but then have it do more than one thing. So, given that there are ten bazillion options for workflow managers, the manager should provide to its user a template example. The template should build and test a container, in this fashion, that is intended to plug into that particular workflow. I would, however, want to see the tests extended. How? Given the workflow, and definitions of inputs and outputs, I am interested to see how we might test interactions between modules along with input and output validation.
Experiment or Data Submission
Given that a PR can be enough to do a deployment, it would be reasonable for a single person to maintain a repository that others change, submit a PR, and the PR updates the deployment. Let’s say when testing happens (we might have data quality checks, or just extraction of metadata) an API endpoint is used to upload it to some repository. Maybe a container with some tag is built using the complete metadata with version according to that timepoint? This is an easy way to use continuous integration to collaboratively build datasets (or collect data), and I’m thinking that the Kaggle-API might be a reasonable first shot (see my comment at that post, we need to discuss the proper endpoints to use!)
Feedback!
Currently, we don’t have a versioning standard for deployment - the project deploys the container to “latest.” How do you think we should go about this? If you would like to ask a question, contribute, or just say hello, please reach out!
Suggested Citation:
Sochat, Vanessa. "The Scientific Filesystem Container Builder." @vsoch (blog), 23 Jun 2018, https://vsoch.github.io/2018/scientific-filesystem-builder/ (accessed 01 Jul 25).