We recently published the Research Software Encyclopedia and also have added several new parsers for obtaining new data, meaning the total collection of curated research software is greater than 1500 entries. In honor of this collection, and of a library I’m working on called CiteLang, I wanted to do a small study to better understand:
- What are the most valuable dependencies in our community, across languages?
- What are the most valuable dependencies in our community, by language?
- What is the credit allocation for each repository?
CiteLang
To step back for a second, let’s talk again about CiteLang. It has many functions - one of them being an ability to assess open source contributions via git, but it’s main purpose is to be a markdown syntax for citing software, meaning that we can:
- Generate basic software credit trees, graphs, and markdown summaries.
- Derive a new, customizable model of credit based on published packages and dependencies.
- Provide a way to cite software in a paper and give credit without needing DOIs.
As a simple example, I can run CiteLang over this markdown file with CiteLang references:
# Summary
Portability and reproducibility of complex software stacks is essential for researchers to perform their work.
High Performance Computing (HPC) environments add another level of complexity, where possibly conflicting
dependencies must co-exist. Although container technologies like Singularity @conda{name=singularity} make
it possible to "bring your own environment," without any form of central strategy to manage containers,
researchers who seek reproducibility via using containers are tasked with managing their own container
collection, often not taking care to ensure that a particular digest or version is used. The reproducibility
of the work is at risk, as they cannot easily install and use containers, nor can they share their software
with others.
Singularity Registry HPC (shpc) @pypi{name=singularity-hpc} is the first of its kind to provide an easy means
for a researcher to add their research software for sharing and collaboration with other researchers to an
existing collection of over 200 popular scientific libraries @github{name=autamus/registry}
@github{name=spack/spack, release=0.17}. The software installs containers as environment modules that are easy
to use and read documentation for, and exposes aliases for commands in the container that the researcher can
add to their pipeline without thinking about complex interactions with a container. The simple addition of an
entry to the registry maintained by shpc comes down to adding a yaml file, and after doing this, another
researcher can easily install the same software, down to the digest, to reproduce the original work.
# References
<!--citelang start-->
<!--citelang end-->
And then run citelang render paper.md to get a nice rendered table alongside your paper! What CiteLang does is find the references in the paper, they look like this:
@conda{name=singularity}
@pypi{name=singularity-hpc}
@github{name=autamus/registry}
@github{name=spack/spack, release=0.17}
Each of the references above is a package manager with a package name and (optionally) a version, and we can load in the metadata for each and then generate a table that you see here that summarizes credit across dependencies. In this model, we give some allocation of credit (default is 50%) to the main work (paper or software) citing the software, and then recursively parse dependencies up to some minimum level of credit to calculate scores. Dependencies shared across libraries are averaged together. The final table represents the credit that you give not only to the top level software, but to all nested dependencies, for the work that you did. And that’s only the basics! CiteLang takes this simple ability to parse references and extends it to automation, graphs, badges, and more! You can read more about CiteLang here.
Publish or perish? How about neither? I just need to keep writing software!
But do you see what is happening above? We aren’t requiring some artificial publication in order to cite software. We are citing it based on its actual usage, as a known dependency to some other software. In a nutshell, we don’t believe that “the traditional academic way” of citing papers makes sense for software, and instead of using DOIs we can use package managers and metadata as a source of truth, and derive the real value of a piece of software based on this ecosystem. This means that as a research software engineer, you can just keep doing what you are already doing, and if someone uses CiteLang to summarize their work, given that your software is published to a package managed you’ll get credit. There are so many cool ideas around this! But let’s start at the beginning. We first want to show how to summarize an ecosystem. That is exactly what we are going to do in this post.
The Research Software Ecosytem
Starting with these curated repositories from a set of scrapers including the Journal of Open Source Software, the HAL Research Software Database, the Research Software NL Dictionary, ROpenSci, and The Molecular Sciences Software Institute, we can do a basic analysis to identify the most used (and thus valued) pieces of software in our ecosystem. My analysis plan was to:
- Start with the current database.
- For each repository, look for requirements files to parse.
- Derive dependency data based on this requirements file.
- Combine and rank to discover the top dependencies!
This of course is limited to the subset of software in our database, and the ability of CiteLang to parse a requirements file. Currently we parse setup.py and requirements.txt (Python), DESCRIPTION (R), go.mod (Go), package.json (npm), and Gemfile (ruby). Based on the breakdown of the languages found in the RSEPedia, this is a reasonable start!
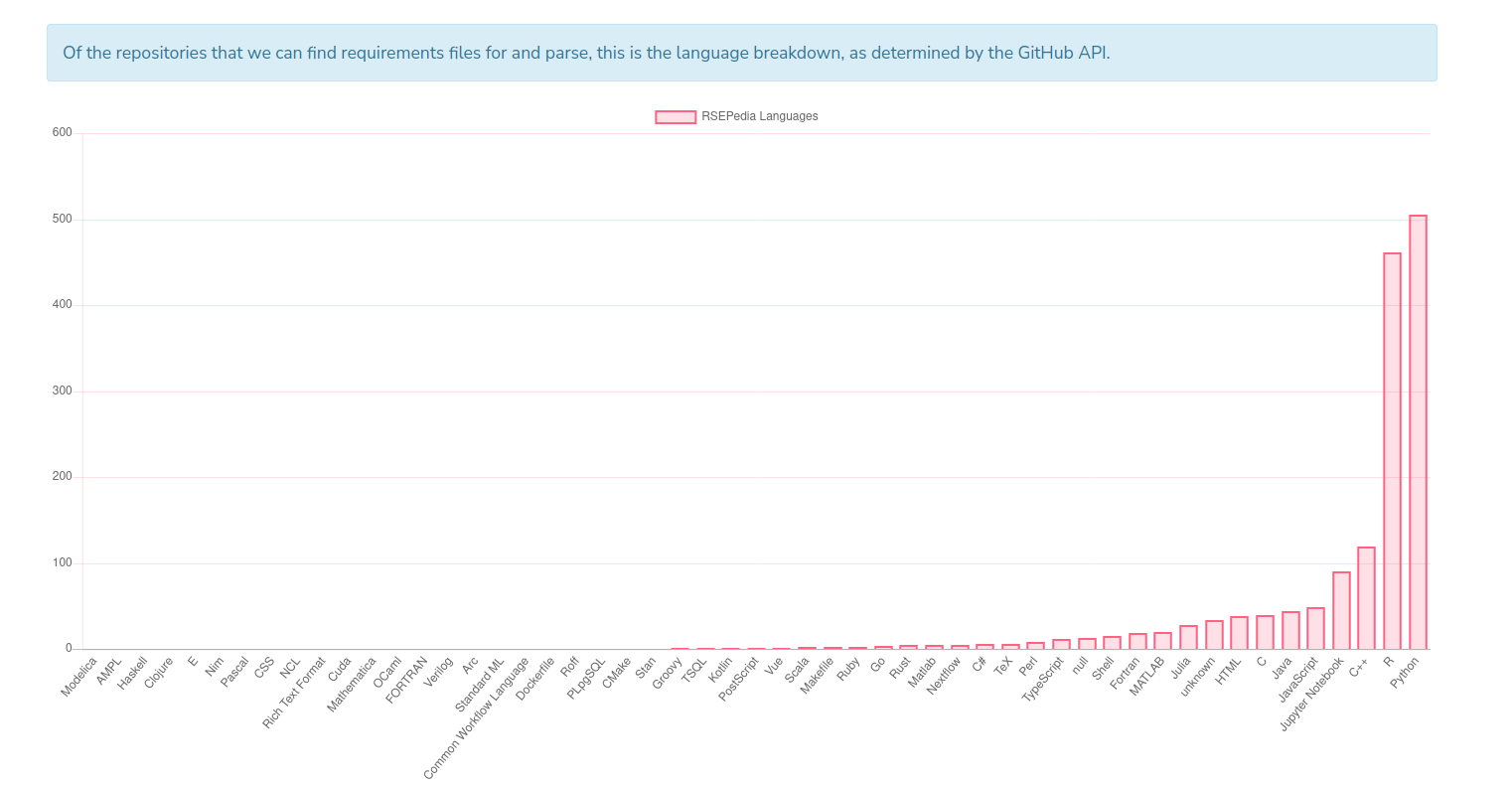
But it’s also kind of sad to see that my favorite languages (Go and Rust) are barely represented in our community. Also, the above should tell you that the R and Python results likely have some meaningful interpretation, but the others not so much, only because we don’t have a big enough sample. So for all of the above steps, for these 1500+ repositories and many languages, I wanted th entire process to be automated, always have potential for easy improvement, and run at some regular interval as new software comes into the Research Software Encyclopedia (also automated) so we can derive changes over time. If you dont’ care to read further:
- View the Research Software Ecosystem
- Check out Languages here
- Results for Dependencies here
- Individual Repositories here
For this first publication of the interface we have the following metrics:
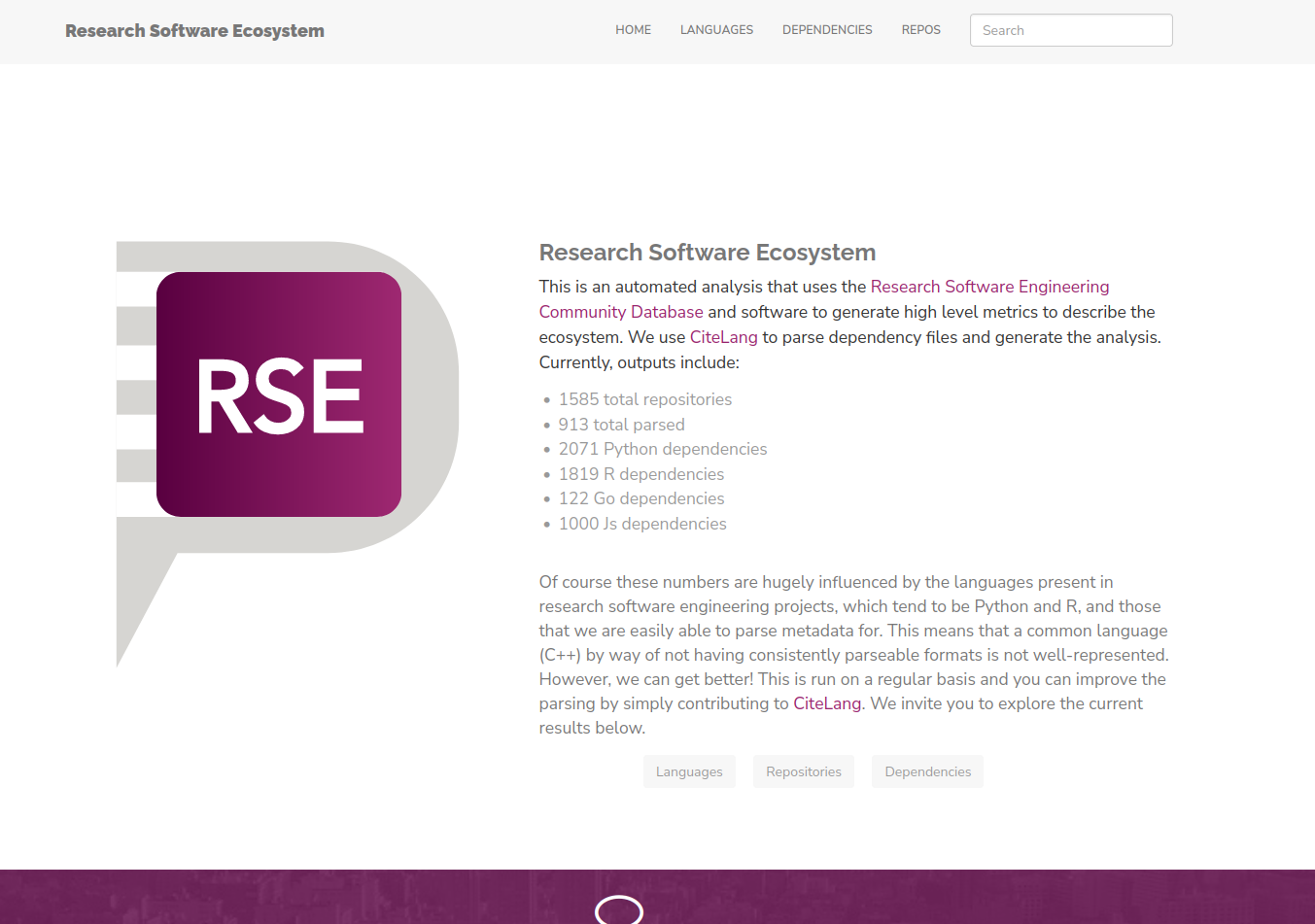
And I’m so excited because a tiny vision I had a few years ago to provide (and use) a community research software database is coming to live! So without further adeiu, I’m just going to jump into the cool results! It will be fun to see how these change over time.
Python
Ladies and gents, dinosaurs and rabbits! Your Python results:
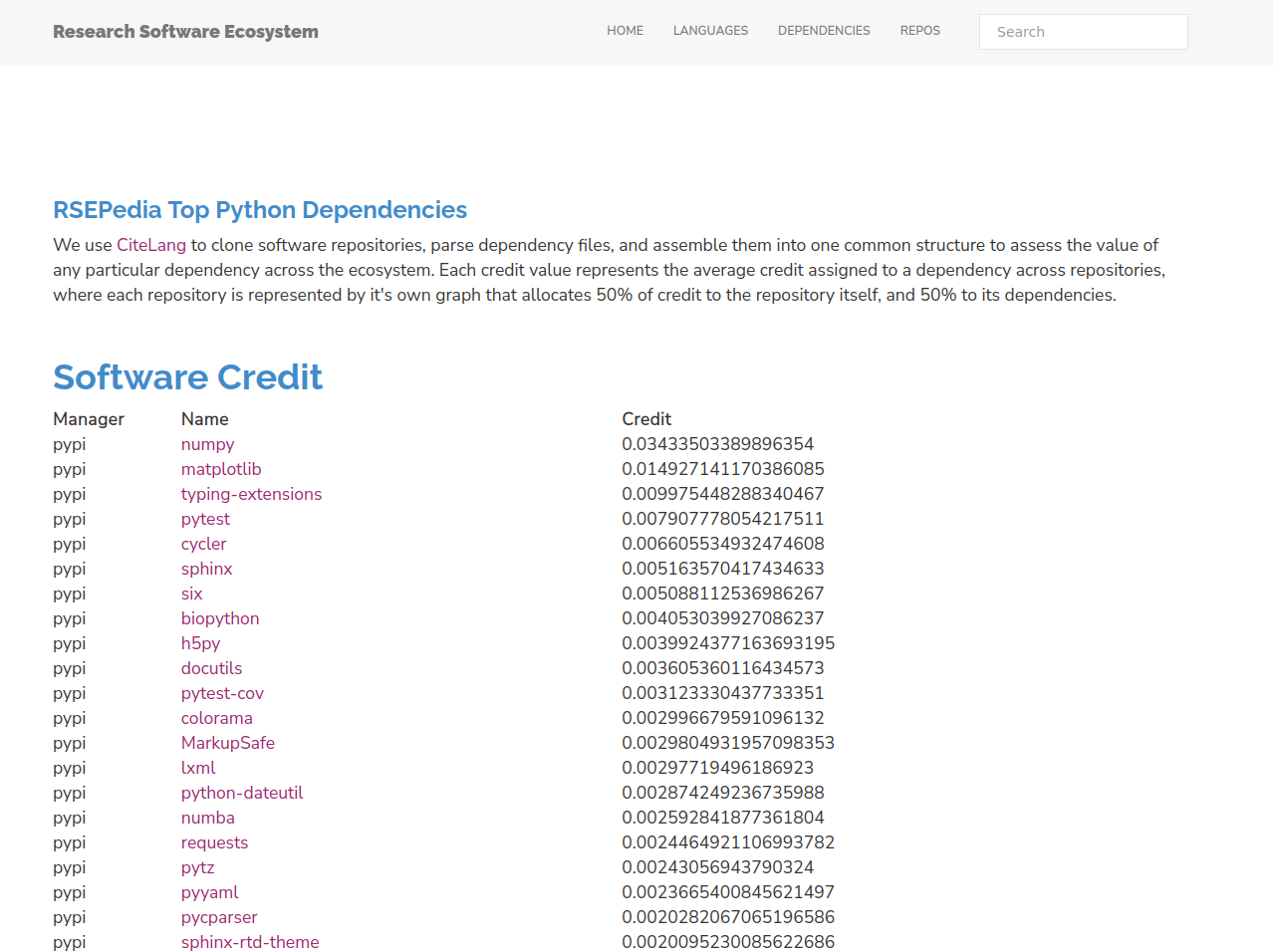
So here is the first awesome insight. Is anyone really surprised to see numpy as the number one library? The credit value here says that the average Python repository is attributing about 3% of credit to numpy, meaning it is a direct or indirect dependency. Let that sink in! Here is the irony - when is the last time you cited numpy? You probably haven’t, because you’ve cited something that uses it. We don’t remember numpy despite the fact that it’s so core to everything that we do.
The fact that the most widely used library is rarely cited is huge evidence for why a manual “write papers and cite DOIs” approach just won’t work for software.
What else do we see in this list? Let me name a few things. First, we can’t be so terrible at remembering to look at or visualize things because matplotlib is second. At least for research software, this is telling us that making plots or charts is important. The next (possibly surprising) result is that documentation and testing is at least represented, and this might be a biased sample because we include repositories that are peer reviewed (e.g., JoSS) and documentation and testing is necessary for that. Given this need for Python, sphinx and pytest come up as leaders to provide that. So here is another nugget of insight:
Some of us are so busy focusing on domain-specific software that we forget the importance of the “less sexy” research software that helps us test, document, view things, or even create simple data structures.
This kind of “base” software has always been what I’ve been most interested in, and ironically what people tell me time and time again “That’s not research software.” Oh really? So something that is entirely powering the research community is not research software? Of course I have my own strong opinions about a taxonomy for research software, but I would encourage those of you who are very dismissive to take a step back and consider what you are really saying.
The next insight is that we see a lot of libraries for data formats (e.g., pyaml, h5py, lxml, and more lower in the list) and this is an attestment to how important being able to read, serialize, and save data is.
The final insight is the fact that requests is high in the list. For those of you not familiar, requests is a library for doing that, making http requests to get content from some webby place. This is an attestment to the fact that our work is increasingly relying on external APIs, automation, or other resources provided on the web.
You can see the full Python results here.
R
I’m less of an R programmer these days, but I think that these results also make sense.
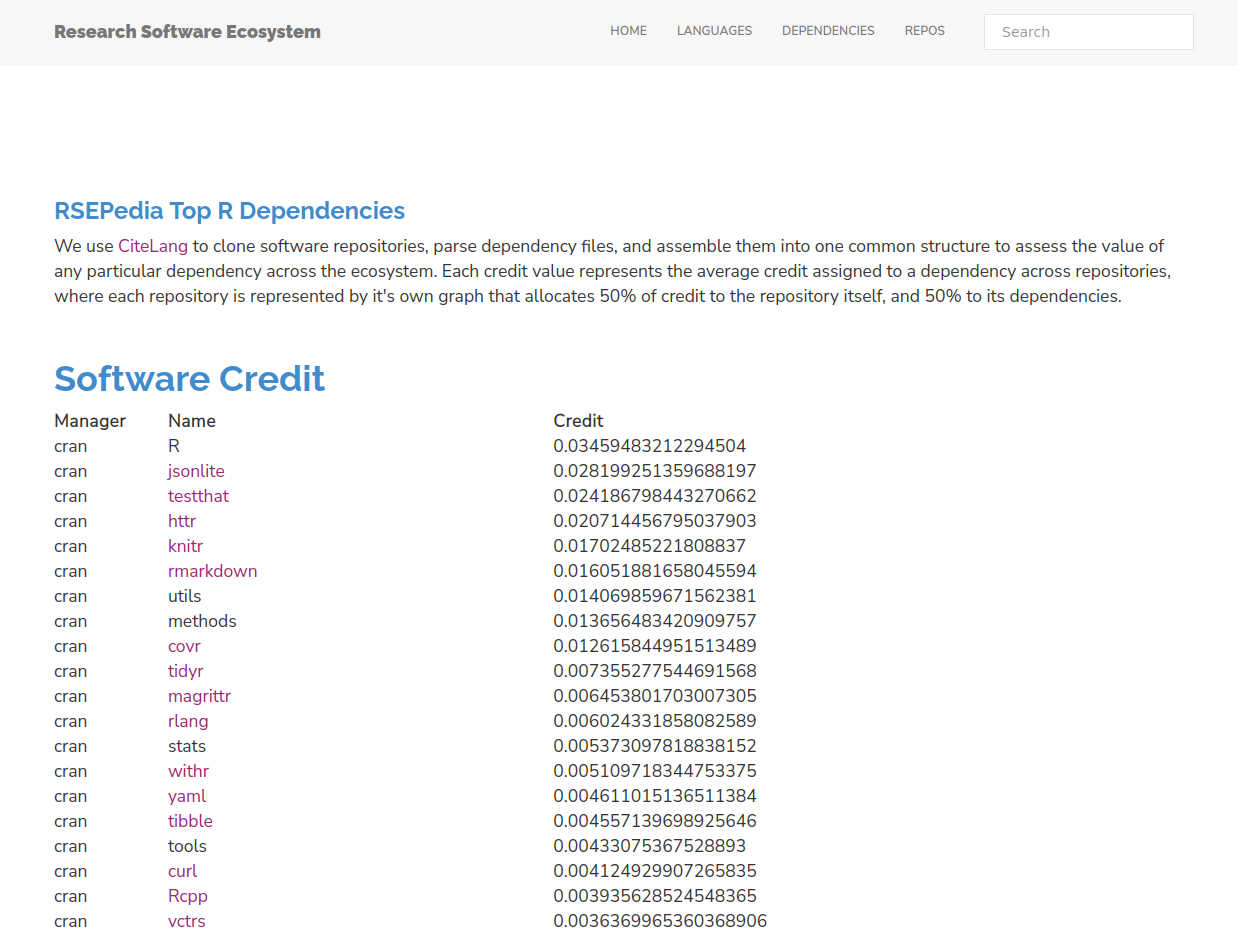
We don’t see any huge leaders in the same way as we see numpy in Python, but not surprisingly the leader package
for the R language is, well, R! I at first thought this was a bug, but actually R DESCRIPTION files that we parse do commonly include a pinned version of R:
Depends: R (>= 3.4.1), TailRank,
...
And so we actually can give credit to the language proper! If you don’t feel this is appropriate, feel free to skip this line and consider the top package jsonlite. This is also why I think json would be represented in Python if it wasn’t part of the standard library. Us research folks - we need our json! Overall I think we see a similar pattern here as we saw with Python. The libraries that float to the top are those that involve data structures (jsonlite, yaml), webby requests or similar (httr, curl), documentation and testing (knitr, rmarkdown) and graphics or visualization. What does this tell us about what is undervalued in research software? Again, it’s not the domain specific libraries, but rather the core stuff that enables those libraries.
You can see the full R results here.
Projects
If you are interested in a specific project in the RSEPedia, we also provide a project-specific table and badge! You can browse projects from here, and here is an example of a badge generated for a project called github.com/ORNL/tx2 (and on GitHub). Without even looking I can tell you we have some machine learning and/or visualization going on here (scikit-learn! umap! pandas! matplotlib)!
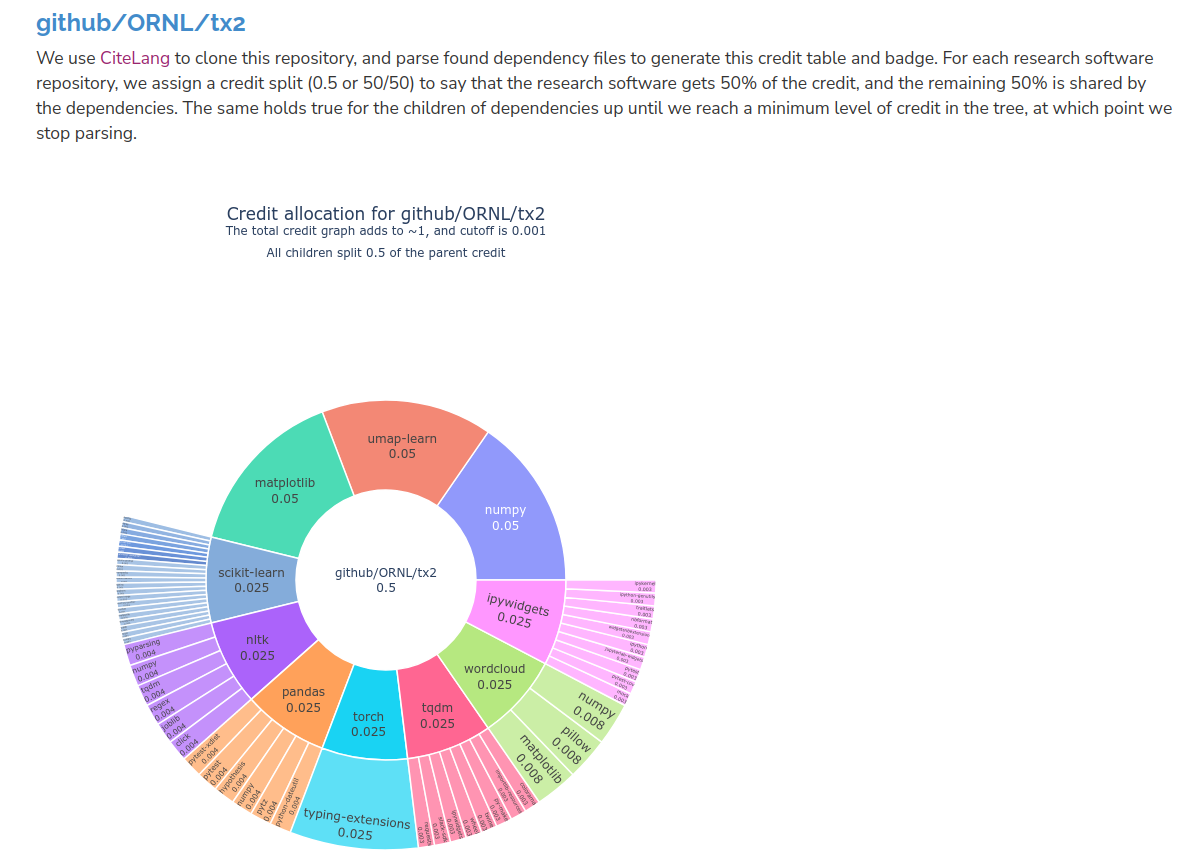
Notice how numpy (as an example) shows up at multiple points in the tree - when we calculate an overall credit, say, for the ecosystem, we take that into account! And we can then peek at the project-specific table and sort of verify that yes, this is a Python ML/visualization project:
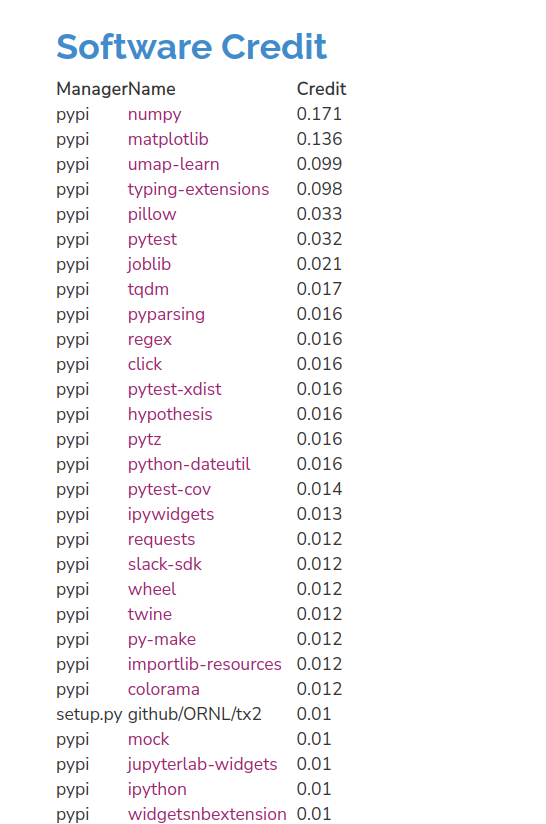
And we see some surprises! Like, the slack-sdk? What? Believe it or not, that is pulled in by tqdm. The project-specific tables (and the description at the top) also give you a better sense of how CiteLang allocates credit. The top level package is given 50%, and then the other 50% is given to all dependencies in the same fashion. We cut off at a value of 0.001, and we do that in case we might be parsing dependencies forever down to some infintesimally small amount.
Finally, every project serves its own raw data
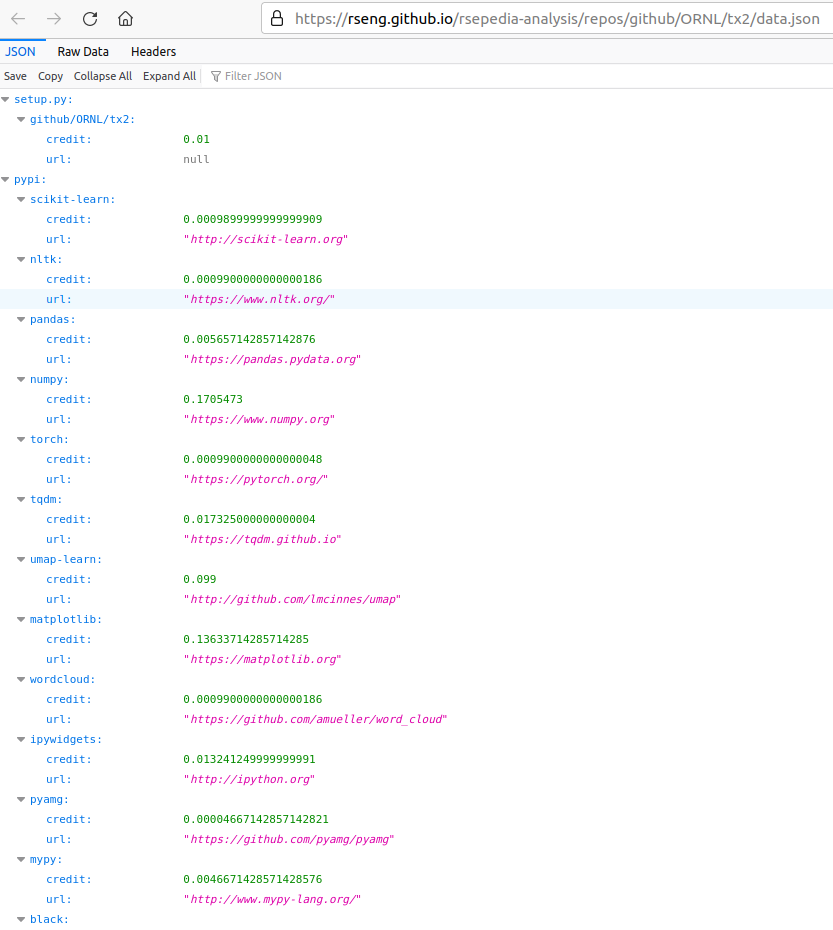
and the site is searchable, because sites should be. 😄️
Discussion
I’m so happy (and a bit relieved, to be honest) to finally be able to show what I’ve been saying for years - that the most valuable software for research, and the software that is driving domain-specific research software, are the unsexy libraries that have to do with data structures, (maybe standards), documentation or testing, and data formats or retrieval. These are the packages that you aren’t going to remember to cite. Also, this set is totally leaving out the software we use on a day to day basis in our CI, which arguably isn’t research software but has done more for the research community than anything I can think of - containers, version control (git), and continuous integration. We’d be a mess without it. We need to be more thankful and aware of this, and for some of y’all that turn down your nose to anything that isn’t a domain-science library, perhaps take a pause. Next, let’s talk about limitations and hopes for the future.
A Living Database
I wouldn’t have been happy with myself to simply publish software at one point in time and call it a day. The Research Software Encyclopedia is updated weekly, and so I’ve designed this analysis to do the same! This means that while we do cache a result for a newly added piece of software, we do continue to grow the analysis as new software is added. And since the tool will always use the newly updated CiteLang, any improvements to the parsers there will be reflected here! And if anyone wants to run the entire thing again (outside of the limit of GitHub Actions) they can clone the repository, nuke the _repos folder, and run the scripts again.
Language Gaps
The biggest gap in the RSEPedia is with respect to what we don’t see. First, despite being a prominent language, we don’t see anything for C++, because there isn’t a package manager with an API to use it. If you have a nifty (or even hacky) idea for how to parse a requirements file, I want to hear it. The RSEPedia has support for spack, but most research-oriented C++ projects are not going to go out of their way to publish their package there, and we get no signal of the package being in spack when we clone the repository. Sppaaaaaack (sorry, it’s a bit of a tic at this point!) 😁️
We also don’t see standard modules or libraries provided within a language. E.g., I can almost guarantee you a ton of Python libraries are importing json, but since it’s not a package manager library we wouldn’t see it. I suspect citelang could come up with a way to derive credit for these libraries by way of abstract syntax trees or just parsing the source code, although I haven’t done this yet because I’m not convinced it’s something people are as interested in. If you want to say thank you for the Python standard library, there is a donate button on their contribution page (or you could contribute code). There is an even deeper level of parsing (at least for Python) that looks at function signatures, and I wrote a library called caliper in early 2021 to do that, and it’s able to generate function databases for Python software of interest. This would be cool to do for some kind of (unrelated) compatibility analysis here, but yes that’s very different.
Parsing Limitation
For all requirements files except for Python, we are forced to do static parsing. While not perfect because bugs can happen for niche cases of someone defining requirements in a weird way, it’s a reasonable start. There is always room for improvement, or adding more static parsers for requirements files I have not considered yet.
However, this is not the case for the Python parsing (either requirements.txt or setup.py)! For Python these results are likely very good because we wrap the pypi package manager install command to derive a list of packages and versions from either a setup.py or requirements.txt. Don’t worry - nothing is installed, we either just parse the requirements file and return the results, or we use the solver against a setup.py to come to an equivalent list. We originally had a static parser (and still use this as a fallback) however I talked to @alecbcs and he had this fantastic idea! Will it likely need updates as time goes on, given the functions are private? Sure. But I’m happy to do that to get the much more accurate listing.
In practice, the only setup.py files that I was not able to parse either had a bug (e.g., trying to read a file that doesn’t exist in the repository) or they were trying to use modules outside of the standard library. For all of the cases of broken-ness, I opened issues on the respective repositories so we might have a better chance at parsing in the future! One detail is that we parse the first requirements file found. For a primary requirements file in the root of the repository, this is the best outcome. However, some repos don’t have a file in the root, and perhaps we find one in a documentation folder instead. Either way, the result represents our best effort at finding and parsing requirements given a cloned repository we don’t know the structure of in advance.
Final Thoughts
Here are my final takeaways:
Publication is not for Research Software
A system of credit that relies on software engineers to do extra manual work (to write papers) is never going to fully capture the ecosystem and give proper credit. It will only capture those that have the time and possibly privilege to take the extra time to write a paper. Publication only makes sense given that a piece of software is paired alongside a robust result, in which case fine, write the paper and also champion the software.
Publication Does not Actually Capture Credit
A system that also only skims the superficial top (the name of one package) and does not dig deep into a dependency tree is also going to miss insights and deserved attributions of credit. As the numpy example shows, nobody is actually citing numpy, but a ton of projects are using it somewhere in their dependency tree, so it deserves a lot of credit.
We Can Do Better
I have a pet peeve. I’m frankly just tired of people writing about credit and attribution but not doing anything about it. We could extend that to other things, but it’s especially an issue for this topic. Ironically they are writing papers and improving their publication record as they write about how publication and research software is a strained process. I may not have solved this problem, but damn at least I’m trying to actually do something about it instead of spurting gas.
I find this idea exciting because there are so many directions you can go with it. When I first designed the idea I imagined a database and online interface where you could essentially connect your GitHub repository, and akin to a builder service, parse your repository on some event and derive a new credit or citation graph. Or you could have some set akin to the RSEPedia that are also updated regularly. And then, by way of having that database, we could do these same queries (that currently I’m doing statically) to say “What are the most important libraries for this language? Across the ecosystem?” or “How has this changed over time?” It would be a true way to derive the value of a library without needing people to publish papers, and totally automated and integrated with package managers, which is where people already should be putting their software. Heck, if someone gave me a cloud and a little bit of funding I’d love to work on this. Are there good reasons or use cases? I don’t know, but maybe.
So what do you think?
Suggested Citation:
Sochat, Vanessa. "The Research Software Ecosystem." @vsoch (blog), 24 Apr 2022, https://vsoch.github.io/2022/rsepedia/ (accessed 03 Jan 26).