Web based experiments must be reproducible, but what does that mean? Typically, a “best effort” might provide code on Github, and pre-register on the open science framework. This is a good start, but can we do better? For example, an open source project near and dear to me, the Experiment Factory places a lot of trust in the creator of the experiments. What is missing?
Testing Reproducible Experiments
The fact that the individual experiments aren’t tested, deployed, and archived is problematic. If I were to install from Github today and build into a container, that’s great. But what if someone else wants to use a previous version of the experiment? With Github it isn’t impossible, but as you move further away from “the thing that I’m actually using” to “scripts run in some place that no longer exists to generate the thing” the probability of success decreases. What would be an improvement? I would want the following things:
- Automation to build a container to deploy the experiment
- Software or functions for working with results
- Testing for the experiment and any analysis software
- A versioned archive of metadata and manifests
- A live preview of the current version
Is this too much to ask?
I don’t think so!
Where does all this stuff go?
It belongs with the experiment. Each experiment should provide its own reproducible testing, development, and sharing, because any packaging and deployment software like the The Experiment Factory cannot take on the burden of these functions at every level.
Who is responsible for doing this?
There is no single person better optimized to write (and provide) code for doing things like processing the results or testing the experiment software than the creator of it. But we must separate the actual functions and experiment from the supporting infrastructure.
The extra work to plug into a build, test, deploy system should not be something that the creator needs to worry about.
This final point was the start of my thinking. While each individual component in the list above might be not too terrible to set up, getting them all together sums to quite a bit of work. It’s not time that everyone has. How can we make this easier? Today I want to discuss some of the steps involved toward this goal. I’m going to be giving a quick overview of the following two experiments:
and then some final thoughts on templates. I challenged my colleagues @earcanal and @tylerburleigh to stick with me as we went on an adventure of Github, CircleCI, R package creation and testing, headless browser drivers, and back!
Example 1: The Breath Counting Task
Earlier this week my colleague @earcanal and I finally chiseled together a complete workflow that I’ve wanted for quite some time. We put together what I would consider the start of my definition for a tested reproducible experiment. Yes, there are improvements to make, but the duct taped thing has taken shape! And it’s a beautiful beastie.
The Workflow Setup
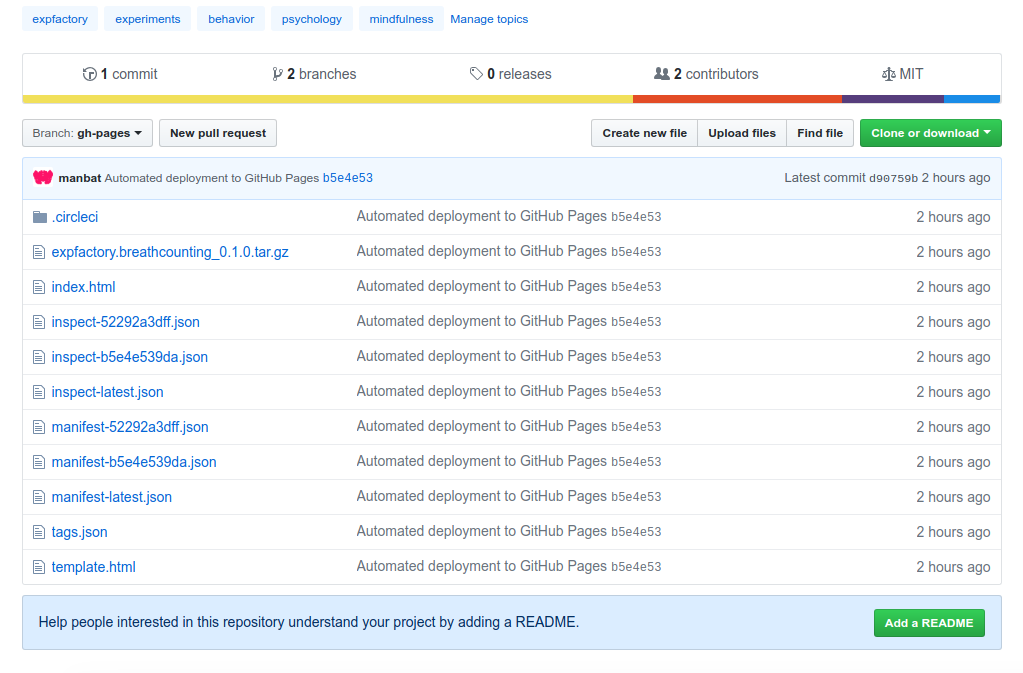
The magic happens with Github and CircleCI, just like this. We start with a Github repository
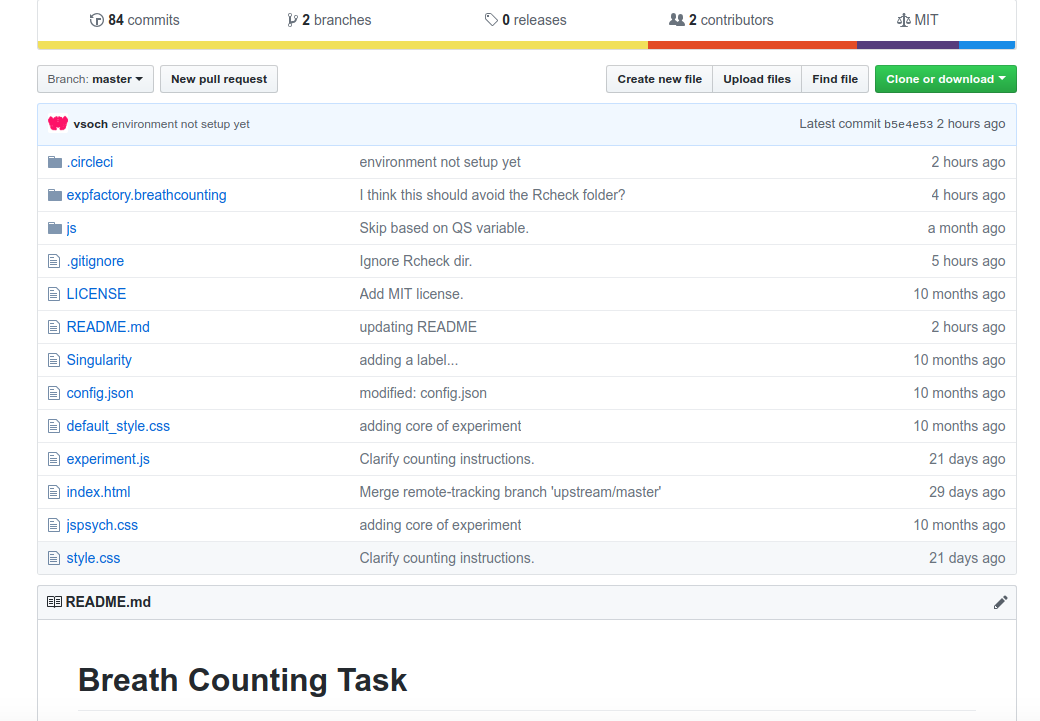
and notice within the repository we have static files for the experiment,
along with a folder (expfactory.breathcounting) that has a package generated by @earcanal
that is functions to work with data generated by this experiment. We then hook it into a continuous integration
service, here is the workflow on CircleCI
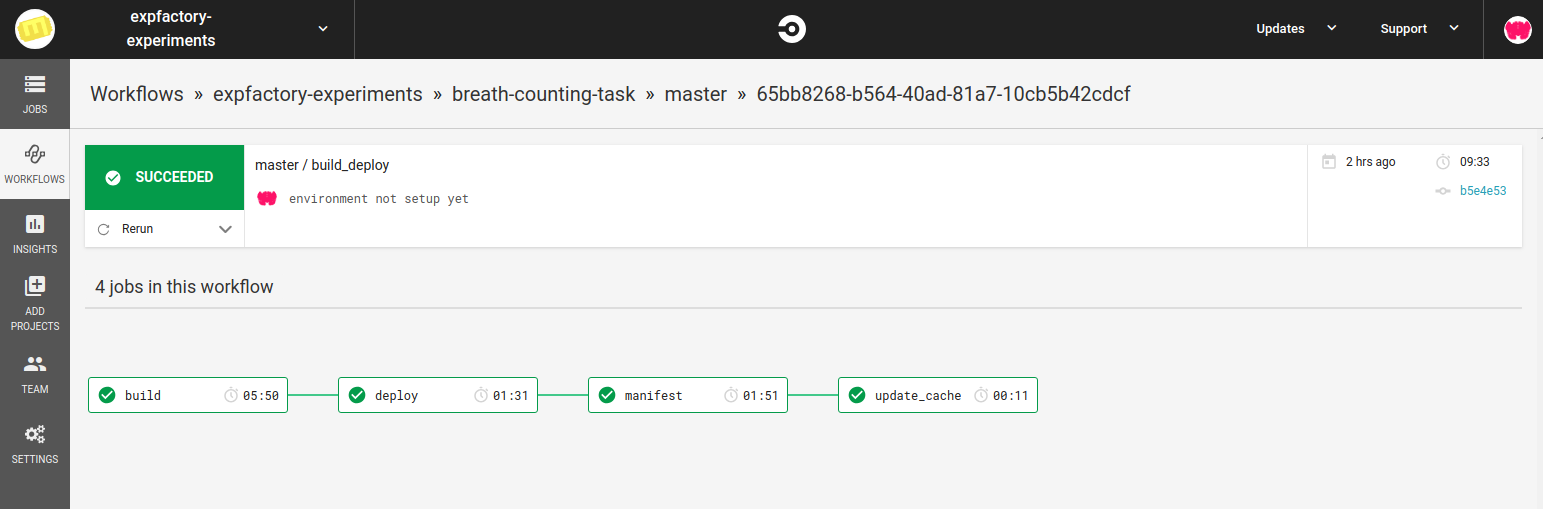
Testing Software
If it’s a pull request, this means that someone is requesting changes to be made to the software. We need to test those changes, and ensure that nothing is broken. This means that we are primarily interested with running the library of tests. Here are tests run by the expfactoryr library to test the Breath Counting Task’s own library. Note that we do this via a Docker Container.
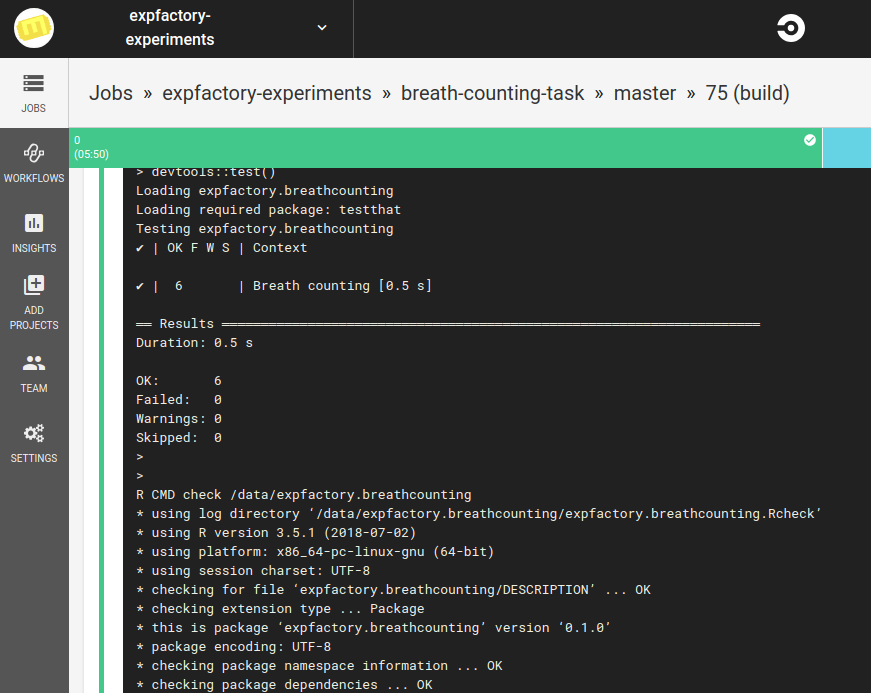
And if the tests pass, we use the expfactory-builder to generate our experiment container!
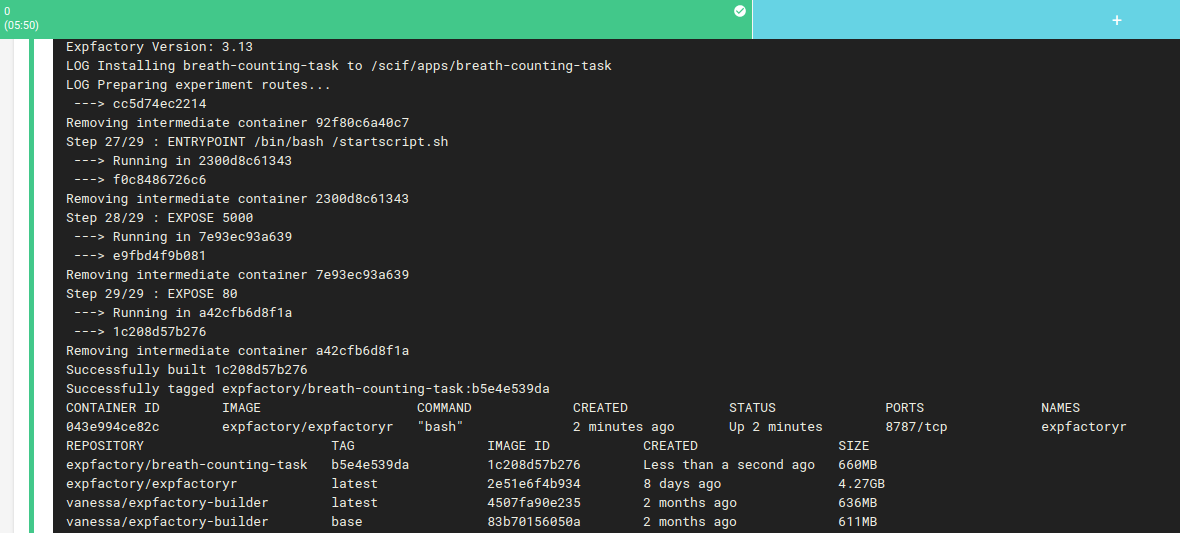
Reproducible Experiment
And then guess what? We push this new container to Docker Hub as expfactory/breath-counting-task, where it’s immediately available for your use.
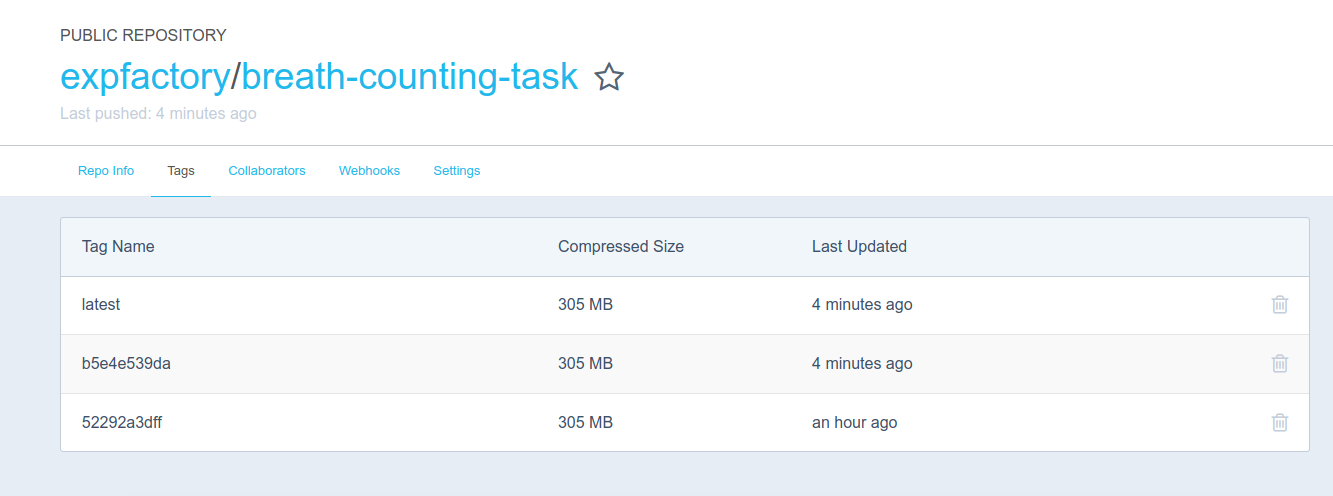
Try it out yourself!
$ docker run -p 80:80 expfactory/breath-counting-task start
and open your browser to http://127.0.0.1 and there it is!
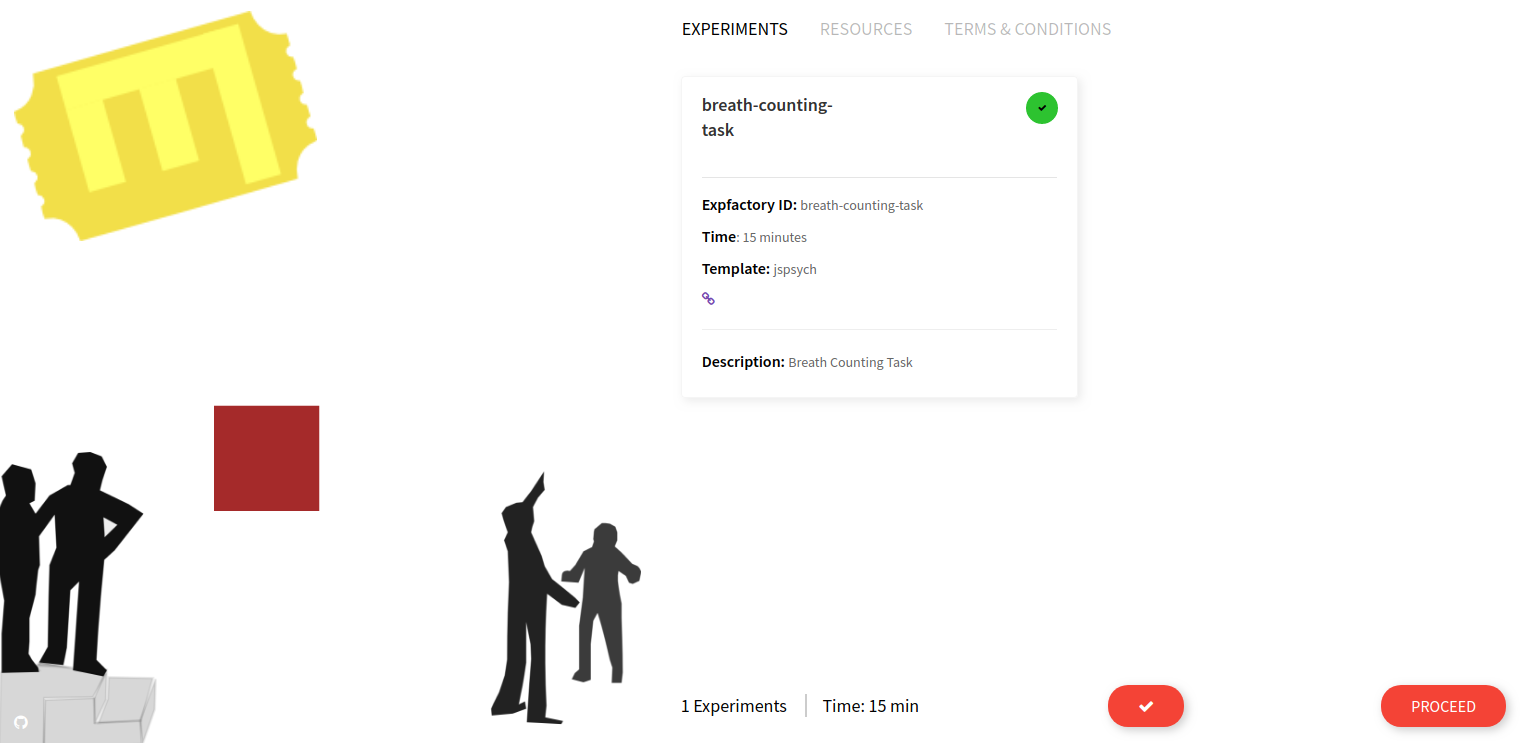
That container is ready to go to be deployed in many different contexts, and since the experiment is provided in the library it’s also ready to go to build into a custom container (if you wanted to serve more than one experiment). You would then build and provide your container, just as we’ve done for the Breathe Counting Task. Cool :)
But that isn’t what I’m most excited about. Remember the package that we built? And what about the container we just used? Complete metadata (the container image manifest and the libraries and versions installed) are generated and sent back to the Github Pages branch.
Example 2: The N-Back 10 Minute Animals
My colleague @tylerburleigh is doing some work to assess if shorter versions of some of our favorite paradigms can be used in place of the longer versions. This one, “N-Back 10 Minute Animals” is one of my favorite because of this:
Those icons are beautiful.
The Workflow Setup
Since I’ve described the workflow with the “Breath Counting Task” I’ll show you this briefly. As we did previously, we also start with a Github repository
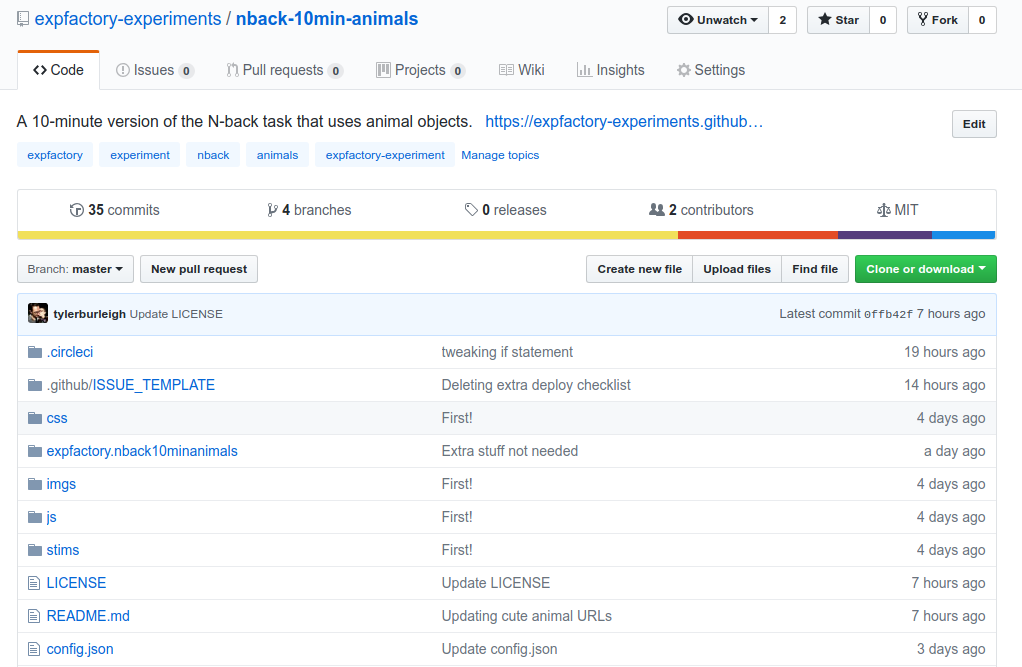
There is equivalently an R package for the researcher to use to process data, and we can see the experiment static files that are rendered on Github Pages for the experiment preview.

And this package is equivalently tested with expfactoryr in the CircleCI workflow. But now we have something different! What about testing the experiment itself? Since this is a JsPsych experiment, we have a robot that then starts and proceeds through the experiment. He primarily ensures that we are able to go from start to end, and there are no 404s or similar. This is done using selenium in Python and Chrome+geckodriver.
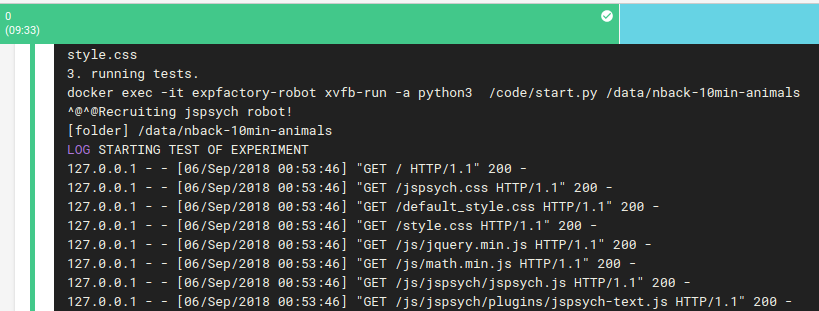
Then if both sets of the tests pass, we use the expfactory-builder to generate our experiment container, just as before, and it gets pushed to Docker Hub, and it’s metadata and manifests generated and put back on Github pages. Here are some quick examples of what is generated:
While it’s not rendered (the master branch is used to serve the task in the repository) there is also a
table index.html file. This means that you can drop the entire gh-pages branch into a web root and get a nice site to explore
container links, and metadata. It looks like this one. And of course to test the task container:
$ docker run -p 80:80 expfactory/nback-10min-animals start
Templates are Important
I probably sound like a broken record, but this simple setup of having code in version control, testing and deploying, and keeping some important outputs or even interactive web content? There are endless use cases! The exciting thing is that while these initial bases can be a lot to create, once you have your template? It is so easy to hand to someone else and they get the same beautiful thing. As a developer, I’ve gone from a mindset of
How do I create a new tool to do the thing
to
How can I made it easier to use already existing things?
And this means that the currency that I want to operate is on the level of webhooks (services) and modular templates to use them. I want to do the hard work of connecting the pipes and making pretty metadata and renderings, and hand it over to the researcher. The researcher can then add what is important to him or her, namely the experiment and analysis. The reproducible products that are essential for his or her success are ready to go. It makes sense, right?
What I’m thinking about
It’s essential that researchers and developers are able to achieve something similar to this with very little work. The above is just a start. From the above, the following needs more work:
Checklists
They work for flying airplanes, and they work here too! Each template needs a checklist of things to do to go from I’m just a template” to “I’m sign, sealed, and delivered!” A first shot at this is adding a Github Issue Template so a user could keep track of their progress filling in the template, and close the issue when finished. If I had an automated way for the user to fork some template repository to “fill in” I would create this issue and direct them to it. Also notice that the template is specific to an “experiment.” I would want to have a selection of different issue templates to provide based on the needs of the template.
Modular Github Pages
It’s fairly straight forward to deploy a single thing to Github pages like a set of manifests or a rendered PDF. But what happens when you put together components that each want their own webby space? They each have their own version of an “index.html” and you either have to combine them or only choose one? I ran into this issue when I realized I couldn’t deploy my table of manifests because the experiment preview was already rendered there. We need to define an organizational structure for the Github Pages content that gives us certainty that each of some set of rendered outputs has a consistent place to live.
Badges
We need to understand what constitutes a template. In the above, we had experiments, a specific testing robot, a testing package in R, and deployment to Docker Hub. I need to be able to look at the Github repository and both identify the template, and the things that make it up. “Badges” can solve this in a fun way, because they come down to categories.
Thanks for stopping by!
If you can’t tell, I’m really excited about this development, and I think it’s going to produce something that is both fun and useful. Over here in dinosaur land there are a few projects underway around this idea, so I’m excited to share those with you when the time is right. :)
Resources
- Github
- Docker Hub
- Experiment Preview
- Metadata and Package
- The Experiment Factory
- Expfactory Experiments
Suggested Citation:
Sochat, Vanessa. "Testing Reproducible Experiments." @vsoch (blog), 06 Sep 2018, https://vsoch.github.io/2018/testing-experiments/ (accessed 01 Jul 25).